SUPPLEMENTARY INFORMATION
|
|
|
- Juuso Hakala
- 6 vuotta sitten
- Katselukertoja:
Transkriptio
1 a b 1
2 c Supplementary Figure 1 Phylogenomic analysis using single gene trees and a concatenated phylogeny. a) Counts of otherwise monophyletic bipartitions containing E. huxleyi from 1563 single protein trees. Black bars represent counts from only those trees in which the branch leading to E. huxleyi and its sister taxon is represented by a bootstrap value of 70 or above. Grey bars represent counts from all trees. b) Concatenated phylogeny of 228 E. huxleyi genes showing the best RAxML topology with support values from 100 bootstrap replicates. Note the poorly resolved placement of the E. huxleyi+guillardia theta clade with respect to the other plastid-containing lineages. c) Counts of otherwise monophyletic bipartitions containing E. huxleyi from single protein trees corresponding to the alignments used in the concatenated tree. Black bars represent counts from only those trees in which the branch leading to E. huxleyi and its sister taxon is represented by a bootstrap value of 70 or above. Grey bars represent counts from all trees. 2
3 Supplementary Figure 2 Asymptotic analysis of the core genome size estimates obtained from comparative genomic hybridization (cgh). The core-genome size from cgh is 12,757 +/- 1,673 genes. From an exponential fit the estimated asymptotic size is ~13,000 (horizontal black line). The unassembled Illunmina comparative approach yielded a core-genome of 17,826 genes (dotted black line) while the assembled reads yielded a core of 14,344 genes (green), see text. Vertical bars indicate ± one standard deviation from 200 random re-samples. The asymptotic estimate is the average from repeated non-linear exponential fits under 500 random permutations of the order of hybridization data and dropping estimates lower than 8,
4 Supplementary Figure 3 The cumulative number of genes in the reference genome not detected in the indicated number of strains (~9000 variable genes are not detected in at least one strain, but all of these genes are detected in the genetic repository of all strains). Illumina reads generated from genomic DNA extracted from 13 different strains, including AWI1516, were mapped to the refined gene model set assembled from the reference genome. Genes were detected using a threshold value equivalent to the coverage of ~50% of the gene length), whereby genes with < 50% coverage with mapped reads were regarded as missing. The total number of missing genes is represented on the Y-axis and the number of strains is indicated along the X-axis. Supplementary Figure 4 The percentage of reference genes detected across multiple strains of E. huxleyi. Illumina reads generated from genomic DNA extracted from13different strains, including AWI1516, were mapped to the reduced gene model set assembled from the reference genome. Genes with > 50% coverage based on mapped reads were called present, and the total number of genes found in each strain was then compared to the 30,569 predicted gene models in the reference genome. 4
5 a. Intron donor site logo Canonical Non-canonical b. Intron acceptor site logo c. Intron length distribution Supplementary Figure 5 Logo images of intron donor and acceptor sites. a, The sequence logo for the E. huxleyi canonical and non-canonical donor sites in the data set with the most frequent nucleotides being CAGGT GCGC and CAGGC GCGC. b, The sequence logo for the acceptor sites in the E. hxuleyi data set with the most frequent nucleotides being GT/GC AGG. c, Intron length distribution where GC donor introns are shown in red and the GC donor introns are in blue. The vertical axis shows the percentage of introns and the horizontal axis, the base pair length of the introns. 5
6 Supplementary Figure 6 Pie charts of the function categories of genes in the pan genome (core and variable sets based on the reference strain CCMP 1516). Colors are coded according to the insert legend, with percentages given in parentheses for the core versus variable genomes. Note that the major difference is the percentage of unknown/orphan genes. 6
7 Supplementary Table 1 Emiliania huxleyi CCMP 1516 reference genome assembly statistics. General assembly Total number of reads Genome size (Mb) Read depth coverage (fold) 10 Scaffold sequence gap (%) 6.98 Scaffold number 6995 Scaffold N50/L50 109/404.8 Kb Contig number Contig N50/L /29.7 Kb Average Properties Gene length (bp) 1718 Transcript length (bp) 1129 Protein length (aa) 346 Exon frequency number per gene 3.65 Exon length (bp) 308 Intron length (bp) 242 Gene density per Mbp 233 Estimating genome completeness: Conserved eukaryotic single copy genes a 97% Core eukaryotic genes mapping approach (CEGMA) b 97% Genes were validated by the following evidence: Total number of gene models 30,569 (100%) Start + stop codons 25,315 (83%) EST support (# of genes / %) 15,642 (51%) RNASeq (# of genes / %) 29,637 (96%) Tiling array (# of genes / %) 23,911 (72%) NR hit (# of genes / %) 21,143 (69%) SwissProt hit (# of genes / %) 19,722 (65%) Pfam hit (# of genes / %) (34%) abased on the 716 highly conserved single-copy eukaryotic genes detailed in the Daphnia pulex genome paper. bbased on the 458 highly conserved single-copy eukaryotic genes from the CEMGA set. 7
8 Supplementary Table 2 Genes putatively identified as encoding enzymes involved in the catabolism of DMSP. tblastn homology searches were performed against the genome using corresponding protein sequences from Halomonas sp. HTNK1. Gene Description Gene Name No. of Orthologs Protein ID Allelic Variant EST evidence e-value Homolog Choline dehydrogenase DddA Y 1.32E-42 Halomonas sp. HTNK Y 2.94E Y 3.29E Y 1.18E Y 5.53E Y 4.89E-10 DMSP lyase DddD Y 1.56E-63 Halomonas sp. HTNK Y 2.75E-55 Enoyl-CoA hydratase AcuK Y 1.51E-31 Halomonas sp. HTNK Y 1.51E Y 8.00E N 1.80E Y 6.28E-08 Iron containing alcohol Pseudomonas sp. J465 dehydrogenase DddB Y 1.79E Y 9.28E-25 Aldehyde dehydrogenase DddC Y 2.10E-25 Halomonas sp. HTNK Y 3.97E Y 1.16E
9 Y 6.58E-20 MMPA-CoA ligase DmdB Y 1.30E-20 Ruegeria pomeroyi Y 3.06E Y 3.06E N 1.75E N 1.75E Y 4.27E Y 1.62E-11 MMPA-CoA dehydrogenase DmdC / Y 1.38E-37 Ruegeria pomeroyi Y 9.47E Y 8.18E-10 MTA-CoA hydratase DmdD N 2.16E-06 Ruegeria pomeroyi 9
10 Supplementary Table 3 Proteins involved in sphingolipid and omega-3- polyunsaturated fatty acid synthesis. Protein Description Functionally Characterized No. of Paralogs Protein ID Core vs. Variable Genome Genes involved in sphingolipid synthesis Serine Palmitoyl Transferase (SPT) yes core Ketosphinganine reductase no core no core Ceramide synthase (LAG1) yes nd Ceramide glucosyltransferase no core Sphingolipid LCB 4-desaturase yes core Sphingolipid LCB 8-desaturase yes core Sphingolipid LCB 9-methyl transferase yes core Genes involved in the biosynthesis of omega-3 LC-PUFAs 12-desaturase no nd no core 15-desaturase no core 6-desaturase no core 9-elongase no core no core 8-desaturase no core 5-desaturase no core 5-elongase no core no core 4-desaturase no core 10
11 Supplementary Table 4 Genes encoding proteins potentially involved in biomineralization processes. Protein Description Family member/ Subfamily/ Complex subunit Number of Paralogs Protein ID Core vs Variable Proton/calcium exchanger CAX core variable core core core nd Cation/calcium exchanger CCX core core Potassium dependent sodium/calcium exchangers NCKX core variable Potassium independent sodium/calcium exchangers nd core core variable core core core core core NCX core Calcium pumps ECA nd Ca 2+/ Mg 2+ -permeable cation channels Transient Receptor Potential TRP core 4 Domain voltage-gated Ca 2+ channels CAV core 11
12 VDC core nd IP3R IPR nd variable Eukaryotic Na + /H + exchanger NAH core core core core core Bacterial Na + /H + exchanger NHA variable core core nd Voltage gated proton channel HVCN nd V-ATPase ATPVA core ATPVA core ATPVB core ATPVC core ATPVC core ATPVC nd ATPVD core ATPVD core ATPVE core ATPVF core 12
13 ATPVG core P type ATPase (PM H + ATPase) variable core core core core H + PPase core Putative Ci Transporters SLC4 Na + independent Cl - /HCO3 - exchangers SLC core AE core core core Carbonic anhydrases Alpha CA core core / nd Beta CA nd nd variable variable Delta CA core core core core variable variable 13
14 Gamma CA variable variable core core 14
15 Table 5 A comparison of the sanger sequenced CCMP 1516 reference genome with the CLC assemblies of three other E. huxleyi strains deeply sequenced using the Illumina platform. Strain Flow Cytometry Haploid Genome Size (Mb) Genome Assembly Size (Mb) Illumina Read Depth Coverage CEGMA % Completeness Ref Genome Coverage (%) Non-aligning sequence a (Mb) Strain Specific Sequence b (Mb) M202 (EH2) 120 +/ (47%) 40.7 (35%) M222 (VAN556) 133 +/ (39%) 21.4 (20%) 92A (CCMP379) 99 +/ (23%) 8.8 (10%) a Non-aligning sequence, the amount of assembled non-gapped sequence with blast identity scores to the CCMP 1516 reference genome < 80% over segments > 100 bp; with percentage of the total strain specific assembly specified in parenthesis. b Strain Specific sequence, the amount of assembled non-gapped sequence with blast identity scores to other strains < 80% over segments > 100 bp; with percentage of the total strain specific assembly specified in parenthesis. c Missing genes are defined as genes with <50% length coverage by mapped reads. 15
16 Supplementary Table 6 Characteristics of E. huxleyi strains used for Illumina sequencing. Strain Location Length in Culture Flow Cytometry Genome Size (Mb) Genome Coverage % Ref Genome Covered b %Unmapped Reads Missing SNPs Genes c AWI1516 a South Pacific ( S W) / ,889 92D English Channel (50,02N 04,22W) / ,925 92E English Channel (49, 52N 6,12W) / ,800 92F English Channel (49,52N 6,12W) 1992 nd nd ,053 B11 Bergen Sea (60, 16N 5,14E) / ,854 B39 Bergen Sea (60 25' 58" N, 5 38' 13" E) / ,100 L Oslo fjord (60 N 11 E) 1959/68/80 nd nd ,623 M217 Bergen Sea (60,16N 5,14E) / ,897 12_1 (CCMP 371) Sargasso Sea (32N 62W) 1987 nd nd ,487 M219 (NZEH) Big Glory Bay, New Zealand / ,536 M202 (EH2) Gt. Barrier Reef, Australia / ,166 92A English Channel / ,785 M222 (VAN556) Vancouver, BC (49,5N 144,40W) / ,852 a AWI1516 * was nominally considered CCMP1516 but maintained at the Alfred Wegener Institute and is not the CCMP culture collection strain used for JGI Sanger sequencing. b Coverage was estimated by direct mapping of reads to the reference genome using the Mapping and Assembly with Quality software package. c Missing genes are operationally defined as genes with less than 50% of their length covered by mapped reads. 16
17 Supplementary Table 7 Assembly statistics of genomic reads from 12 different E. huxleyi strains using the CLC Genomics Workbench. Strain Total Reads Total bp % GC N a n:n50 b N50 c (bp) Max d (bp) Assembly Size (Mb) Completeness CEGMA (%) e AWI ,260, D 26,153,077 1,987,633, E 27,029,626 2,054,251, F 26,501,216 2,676,622, B11 23,074,156 2,330,489, B39 22,759,741 2,298,733, , L 22,905,701 1,740,833, M217 24,012,306 2,425,242, (CCMP371) 26,604,262 2,021,923, M219 (NZEH) 50,355,481 4,462,328, M202 (EH2) 386,571,098 38,657,109, A 344,534,050 34,453,405, M222 (Van556) 352,924,546 35,292,454, total number of contigs b contig number of the median contig length a c median contig length d maximum contig length ee percent completeness based on conserved eukaryote genes mapping approach based on criteria using an alignment coverage >50% of the gene length 17
18 Supplementary Table 8 E. huxlei genes likely transferred from bacteria or viruses. Protein ID Gene Description Bacterial/Viral homologs e-value HisI Phosphoribosyl-AMP cyclohydrolase Verrucomicrobia (Coraliomargarita akajimensis) 8.00E TrxB Thioredoxin reductase PaaY Carbonic anhydrases/acetyltransferases, isoleucine patch superfamily Uncharacterized conserved protein CaiD Enoyl-CoA hydratase/carnithine racemase DdpA ABC-type dipeptide transport system, periplasmic component HisC Histidinol-phosphate/aromatic aminotransferase and cobyric acid decarboxylase Cyanobacteria (Synechococcus sp., Prochlorococcus marinus) 2.00E-101 Verrucomicrobia (Coraliomargarita akajimensis, Akkermansia muciniphila) 9.00E-45 Cyanobacteria (Synechococcus sp., Prochlorococcus marinus) 6.00E-137 Actinobacteria (Frankia sp., Acidimicrobium ferrooxidans) 5.00E-28 Fusobacteria (Fusobacterium nucleatum) 2.00E-57 Gammaproteobacteria (Saccharophagus degradans) 4.00E DraG ADP-ribosylglycohydrolase Epsilonproteobacteria (Arcobacter nitrofigilis) 2.00E RacX Aspartate racemase Betaproteobacteria (Burkholderia cepacia, Burkholderia ambifaria) 6.00E unknown Betaproteobacteria (Bordetella avium, Bordetella bronchiseptica, Bordetella petrii, Achromobacter xylosoxidans) 4.00E AslA Arylsulfatase A and related enzymes Verrucomicrobia (Coraliomargarita akajimensis) 4.00E unknown Actinobacteria (Mycobacterium smegmatis) 3.00E unknown Alphaproteobacteria (Jannaschia sp.) 4.00E Predicted CoA-binding protein Uncharacterized conserved protein Predicted integral membrane protein linked to a cation pump Firmicutes (Clostridium perfringens, Clostridium botulinum, Clostridium cellulovorans) 3.00E-19 Actinobacteria (Geodermatophilus obscurus, Beutenbergia cavernae) 2.00E-13 Alphaproteobacteria (Agrobacterium tumefaciens) 1.00E
19 unknown Bacteroidetes (Paludibacter propionicigenes) 7.00E Predicted esterase Deltaproteobacteria (Haliangium ochraceum) 2.00E HemL Glutamate-1-semialdehyde aminotransferase Alphaproteobacteria (Methylobacterium radiotolerans, Methylobacterium nodulans, Methylobacterium sp.) 7.00E MhpC Predicted hydrolases or acyltransferases Betaproteobacteria (Burkholderia xenovoran) 3.00E TrkA Predicted flavoprotein involved in K+ transport Actinobacteria (Mycobacterium smegmatis) 6.00E Hfi Hydroxypyruvate isomerase Bacteroidetes (Spirosoma linguale, Zunongwangia profunda, Maribacter sp.) 4.00E CysC Adenylylsulfate kinase and related kinases Cyanobacteria (Synechococcus sp.) 6.00E Predicted amino acid aldolase or racemase Acidobacteria (Solibacter usitatus, Candidatus Koribacter versatilis ) 1.00E PyrF Orotidine-5'-phosphate decarboxylase Alphaproteobacteria (Azospirillum sp.) 8.00E Icc Predicted phosphohydrolases Gammaproteobacteria (Legionella pneumophila) 9.00E CysK Cysteine synthase Firmicutes (Clostridium difficile) 4.00E Era GTPase PurC Phosphoribosylaminoimidazolesuccinocarboxamide synthase GlmS Glucosamine 6-phosphate synthetase, Bacteroidetes (Dyadobacter fermentans, Leadbetterella byssophila) 3.00E-72 Gammaproteobacteria (Ferrimonas balearica, Vibrio cholerae) 8.00E-149 Bacteroidetes (Salinibacter ruber, Rhodothermus marinus) 4.00E unknown Betaproteobacteria (Variovorax paradoxus) 5.00E PutA NAD-dependent aldehyde dehydrogenases MntH Mn 2+ and Fe 2+ transporters of the NRAMP family Protein involved in biosynthesis of mitomycin antibiotics/polyketide fumonisin Actinobacteria (Streptomyces scabiei, Mycobacterium abscessus) 4.00E-157 Bacteroidetes (Rhodothermus marinus, Spirosoma linguale) 2.00E-22 Deltaproteobacteria (Sorangium cellulosum, Myxococcus xanthus) 8.00E CaiA Acyl-CoA dehydrogenases Alphaproteobacteria (Parvularcula bermudensis) 8.00E Rpe Pentose-5-phosphate-3-epimerase Cyanobacteria (Prochlorococcus marinus) 5.00E Short-chain alcohol dehydrogenase of unknown specificity Alphaproteobacteria (Roseobacter denitrificans, Maricaulis maris, Hirschia baltica) 2.00E
20 SpsE Sialic acid synthase Bacteroidetes (Croceibacter atlanticus) 6.00E PdxK Pyridoxal/pyridoxine/pyridoxamine kinase Gammaproteobacteria (Dickeya dadantii) 8.00E AceE Pyruvate dehydrogenase complex, dehydrogenase (E1) component CaiA Acyl-CoA dehydrogenases Alphaproteobacteria (Sphingomonas wittichii, Caulobacter sp., Bradyrhizobium sp., Acidiphilium cryptum) 0 Gammaproteobacteria (Pseudomonas syringae, Pseudomonas putida) 9.00E unknown Bacteroidetes (Pedobacter heparinus) 5.00E Predicted acetamidase/formamidase CaiA Acyl-CoA dehydrogenases Alphaproteobacteria (Sphingomonas wittichii, Rhizobium leguminosarum) 3.00E-46 Gammaproteobacteria (Pseudomonas syringae, Pseudomonas entomophila) 2.00E Sphingomonas wittichii) 2.00E-41 Acd 1-aminocyclopropane-1-carboxylate deaminase Deltaproteobacteria (Desulfovibrio desulfuricans) 2.00E WcaA Glycosyltransferases involved in cell wall Deltaproteobacteria (Desulfovibrio desulfuricans) biogenesis 3.00E-28 Alphaproteobacteria (Rhizobium leguminosarum, Predicted acetamidase/formamidase ELO, GNS1/SUR4 family Emiliania huxleyi virus E Dihydroceramide desaturase (Dsd1, delta-4) Emiliania huxleyi virus E Phosphate permease Emiliania huxleyi virus E Lipocalin-like Emiliania huxleyi virus E ERG3, Sterol desaturase Emiliania huxleyi virus E Hypothetical protein Emiliania huxleyi virus E Putative DNA cytosine methylase Enterobacteria phages 5.00E Hypothetical protein Emiliania huxleyi virus E Lipid phosphate phosphatase (PAP2 superfamily) Emiliania huxleyi virus E Fatty acid desaturase (Aco1, delta-9) Emiliania huxleyi virus E Hypothetical protein Emiliania huxleyi virus E Hypothetical protein Emiliania huxleyi virus E Dihydroceramide synthase Emiliania huxleyi virus E
21 Hypothetical protein Emiliania huxleyi virus E-08 DNA repair/recombination protein pif1-like with HRDC domain Emiliania huxleyi virus E ERG3, Sterol desaturase Emiliania huxleyi virus E MFS_1, Major Facilitator Superfamily Emiliania huxleyi virus E Tmk, Thymidylate kinase Emiliania huxleyi virus E Glycosyl transferase family 8-like protein Emiliania huxleyi virus E YqaJ viral recombinase family PBCV-1, OtV5 5.00E Hypothetical protein NCLDVs (Mimivirus, PBCVs, ASCV1) 4.00E ATP-dependent DNA ligase Emiliania huxleyi virus E Sec14p-like lipid-binding domain Emiliania huxleyi virus E Hypothetical protein Emiliania huxleyi virus E Serine palmitoyltransferase Emiliania huxleyi virus E Hypothetical protein Emiliania huxleyi virus E Hypothetical protein Emiliania huxleyi virus E OG-FeII_Oxy domain-containing protein Synechococcus phage Syn9 6.00E Hypothetical protein Emiliania huxleyi virus E Hypothetical protein Emiliania huxleyi virus E Methyltransferase Emiliania huxleyi virus E-22 NCLDVs (Mimivirus, EsV1, OtV5, PBCVs, SSL2, DNA or RNA helicases of superfamily II ASCV1) 2.00E Hypothetical protein Emiliania huxleyi virus E Formamidopyrimidine-DNA glycosylase Mimivirus 4.00E
22 Supplementary Table 9 Classification of the repeat content of the E. huxleyi genome. Class Category Family coverage (bp) % of Genome Class LTR RT Ty1/Copia Ty3/Gypsy DIRS other* Non-LTR RT LINE Class TIR Harbinger hat PiggyBac Sola Tc1/Mariner MITE** TIR** Helitron Helitron rdna-related*** >80% rdna coverage % rdna coverage % rdna coverage NoCat*** >50% TR coverage <50% TR coverage Host gene*** Tandem repeats and low complexity regions In dispersed repeats Not in dispersed repeats Total ~84Mb ~64% * Family not clearly determined. ** Only terminal inverted repeats were detected. *** Only NoCat and rdna-related with at least 10 copies and host genes with at least 5 copies are counted. Dispersed repeats comprise all above classes. The total repeat content is computed by adding the subtotal of each class (first line of each class) except for "Tandem repeats and low complexity regions" where only the exclusive contribution "Not in dispersed repeats" has to be added. 22
23 Supplementary Table 10 Genes involved in the protection and acclimation to high light. Protein Description Protein ID Core/Variable Light harvesting machinery and photoreceptors UVR8 Photoreceptor nd Blue light cryptochrome variable Blue light cryptochrome core Blue light cryptochrome core Blue light cryptochrome core Blue light cryptochrome core Related to blue light cryptochrome variable Related to blue light cryptochrome variable Putative PAS domain sensor hybrid histidine kinase; phytochrome domain, homology to PHYA red or far red photoreceptor core Similarity to blue light receptors but only has the PAS domain variable variable nd nd nd bd core core core nd nd Phototropin nd Phytochrome-like protein similar to phytochrome core Antioxidants and proteins involved in non-photochemical quenching Peroxiredoxin core core Peroxiredoxin-2E, chlorophastic nd core Thiol peroxidase core Violaxanthin de-epoxidase core Violaxanthin de-epoxidase core Zeaxanthin epoxidase core core 11 kda protein of photosystem II, psbz, critical role in NPQ core Alternative Oxidase variable variable 23
24 Ascorbate peroxidase variable core core variable Glutathione peroxidase core core core core core Catalase core core Superoxide dismutase variable core core core Degradation, assembly and repair of PSII Components Photosystem II stability/assembly factor HCF core Photosystem II stability/assembly factor HCF core Photosystem II stability/assembly factor HCF variable Photosystem II D1 protease core Carboxy-terminal processing protease core core core nd core FtsH cell division protein core FtsH core core High light response proteins High light inducible protein core core 24
25 Supplementary Table 11 Table of 68 Light harvesting complex proteins in the E. huxleyi genome Protein ID Allelic Variant Annotation name Core or Variable Genome Group on Tree Proteomics Detection Lhcf 2 core Red Y Lhcf 3 core Red Y Lhcf 4 core Not resolved N Lhcf 5 core Red Y Lhcf 6 core Chlorophyll a/c group II Y Lhcf 7 variable Chlorophyll a/c group I Y Lhcf 8 variable Chlorophyll a/c group I N Lhcf 9 variable Chlorophyll a/c group I N Lhcf 10 core Chlorophyll a/c group I Y Lhcf 11 core Chlorophyll a/c group I Y Lhcf 12 core Chlorophyll a/c group I N Lhcf 13 core Chlorophyll a/c group I N Lhcf 14 core Chlorophyll a/c group I Y Lhcf 15 core Chlorophyll a/c group I Y Lhcf 16 core Chlorophyll a/c group I Y Lhcf 17 variable Chlorophyll a/c group I N Lhcf 18 nd Chlorophyll a/c group I Y Lhcf 19 variable Chlorophyll a/c group I Y Lhcf 20 variable Chlorophyll a/c group I N Lhcf 21 core Chlorophyll a/c group I N Lhcf 22 core Chlorophyll a/c group I Y Lhcf 23 core Chlorophyll a/c group I N Lhcf 24 core Chlorophyll a/c group I Y Lhcf 25 core Chlorophyll a/c group II Y Lhcf 26 variable Chlorophyll a/c group II Y Lhcf 27 core Chlorophyll a/c group II Y Lhcf 28 core Chlorophyll a/c group II Y Lhcf 29 variable Chlorophyll a/c group II N Lhcf 30 variable Chlorophyll a/c group II Y Lhcf 31 core Chlorophyll a/c group II N Lhcf 32 core Chlorophyll a/c group II Y Lhcf 33 core Chlorophyll a/c group II Y Lhcf 34 core LI818 N Lhcf 35 core LI818 Y Lhcf 36 variable LI818 N Lhcf 36 nd LI818 N * Lhcf 37 core LI818 Y Lhcf 38 variable LI818 N Lhcf 39 variable LI818 N * Lhcf 41 core LI818 N 25
26 460117* Lhcf 42 core LI818 N * Lhcf 43 core LI818 N Lhcf 44 core LhcZ N Lhcf 45 core LhcZ N Lhcf 46 core LhcZ Y Lhcf 47 core LhcZ Y Lhcf 48 core LhcZ Y Lhcf 49 core LhcZ N Lhcf 50 core Red Y Lhcf 51 core LI818 Y * Lhcf 52 core LI818 Y Lhcf 53 core LhcZ N Lhcf 54 core Lhcz N Lhcf 55 core LI818 Y Lhcf 56 core Chlorophyll a/c group I N Lhcf 57 core Chlorophyll a/c group I N Lhcf 58 core Chlorophyll a/c group I Y Lhcf 60 variable Chlorophyll a/c group I Y Lhcf 62 variable Chlorophyll a/c group II Y Lhcf 63 variable Chlorophyll a/c group II Y Lhcf 64 core Chlorophyll a/c group II Y Lhcf 65 core Chlorophyll a/c group II Y Lhcf 66 core Chlorophyll a/c group II Y Lhcf 67 core Chlorophyll a/c group II N Lhcf 68 variable Chlorophyll a/c group II N Lhcf 69 core Red Y Lhcf 70 core Red Y Lhcf 71 nd Not resolved N * denotes the detection of a CO 2 enhancer element 26
27 Supplementary Table 12 Genes encoding proteins putatively involved in phosphate metabolism. Protein Description No. of Paralogs Protein ID Core vs. Variable Genome Transporters General phosphate transporter nd variable core core variable variable variable variable core core High affinity phosphate transporter core variable core Phosphate metabolism Alkaline phosphatse variable core core Alkaline phosphatase domain containing protein core nd 5' nucleotidase core 5' nucleotidase core 5' nucleotidase core 5' nucleotidase variable 5' nucleotidase core Cyclic GMP phosphodiesterase variable Purple acid phosphatase-like protein/phytase core nd 62875/78521 core /67476 nd Inorganic pyrophosphatase core core / core Acidocalcisomal pyrophosphatase core core Nudix hydrolase nd 27
28 45054 core core core Evidence of Phosphonate Metabolism HAD superfamily hydrolase core Haloacid dehalogenase-like hydrolase nd core Carboxyvinyl-carboxyphosphonate phosphorylmutase core variable core ABC transporter with homology to phosphonate transporter core core core core Regulation Two component response regulator nd core Homeobox transcription factor core nd Cyclin dependent kinase type C core Protein kinase nd core variable Phospholipid Replacement Sulfolipid biosynthesis protein core Sulfolipid synthetase core Phospholipid/glycerol acyltransferase core nd N-acetyltransferase 12 isoform core nd 28
29 Supplementary Table 13 Genes encoding proteins putatively involved in nitrogen metabolism. Name Protein ID Core or Variable Genome Nitrogen Metabolism Glutamate synthase core Glutamate synthase core Glutamate synthase nd Glutamate synthase core Glutamate synthase nd Glutamine synthetase core Glutamine synthetase core Glutamine synthetase nd Glutamine synthetase core Glutamine synthetase core Glutamine dehydrogenase core Glutamine dehydrogenase core Inorganic Nitrogen Metabolism Nitrogen fixation protein core Nitrogen fixation protein core Nitrite reductase core Copper nitrite reductase 69470/70548/70565 variable Nitrite transporter nd Nitrite transporter core Nitrite transporter core Nitrite transporter core Nitrite transporter core High affinity nitrate transporter core High affinity nitrate transporter core Nitrate transporter core Nitrate transporter core Nitrate transporter variable Nitrate transporter core Nitrate transporter core Nitrate transporter variable Nitric oxide synthase nd flavohemoglobin nd Cyanate Cyanate lyase nd Cyanate lyase core Cyante transporter ACCT9561-J03.xld-F nd Cyante transporter AWCG1205-H03.xld-F nd Nitriliase core Nitriliase core Nitriliase core Nitriliase core Nitriliase core 29
30 Nitriliase variable Urea Urea transporter core Urea transporter core Urea transporter core Urea transporter variable Urea transporter variable Urea transporter nd Arginase nd Urease nd Carbamoyl phosphate synthetase-iii core Carbamoyl phosphate synthetase-ii core Ornithine transcarbamyolase core Ornithine transcarbamyolase nd Argininosuccinic acid synthetase nd Argininosuccinic acid synthetase core Argininosuccinate lyase core Argininosuccinate lyase core Argininosuccinate lyase nd Ammonium transporter nd Ammonium transporter nd Ammonium transporter variable Ammonium transporter nd Ammonium transporter core Ammonium transporter variable Ammonium transporter variable Ammonium transporter core Ammonium transporter core Ammonium transporter variable Ammonium transporter variable Ammonium transporter nd Ammonium transporter core Ammonium transporter core Ammonium transporter core Ammonium transporter core Ammonium transporter variable Ammonium transporter core Ammonium transporter variable Ammonium transporter variable Other Phenylalanine/Histidine ammonia lyase variable Glutaminase core Glutaminase core Glutaminase nd Glutaminase core Hypoxanthine phosphoribosyltransferase core Polyamine transporter nd Polyamine transporter nd Polyamine transporter nd 30
31 Polyamine transporter nd Polyamine transporter variable Polyamine transporter nd Polyamine transporter nd Polyamine transporter nd Polyamine transporter nd Polyamine transporter nd Polyamine transporter nd Polyamine transporter core Polyamine transporter variable Polyamine transporter nd Polyamine transporter core Polyamine transporter variable Polyamine transporter core Polyamine transporter nd Polyamine transporter nd Polyamine transporter core Polyamine transporter core Polyamine transporter core Polyamine transporter nd Polyamine transporter core Dipeptidase plasmid membrane bound variable Dipeptidase plasmid membrane bound core Dipeptidase plasmid membrane bound nd Dipeptidase plasmid membrane bound nd Amine oxidase core Amine oxidase core Amine oxidase copper core Amine oxidase copper core Amine oxidase copper core Amine oxidase copper variable D-amino acid oxidase core L-amino acid oxidase variable FAD amine oxidase variable FAD amine oxidase variable FAD amine oxidase core Polyamine oxidase core Chinitase core Chitinase core Chitinase core Chitinase variable Chitinase core Chitinase nd Chitinase core Chitinase variable Amidase variable Amidase variable Amidase variable Amidase variable 31
32 Amidase variable Acetamidase/Formamidase variable Acetamidase/Formamidase variable Acetamidase/Formamidase core Acetamidase/Formamidase core Acetamidase/Formamidase variable Acetamidase/Formamidase variable Acetamidase/Formamidase variable Acetamidase/Formamidase variable Acetamidase/Formamidase core Nitrile hydratase beta subunit variable Nitrile hydratase alpha subuni nd Nitrile hydratase alpha subuni nd Nitric oxide reductase Present in unassembled reads 32
33 Supplementary Table 14 Genes encoding putative selenoproteins. Selenoprotein Gene name Protein ID Allelic Variant Dithiodisulfide oxidoreductase DSBA Dithiodisulfide oxidoreductase DSBA Dithiodisulfide oxidoreductase DSBA Dithiodisulfide oxidoreductase DSBA Dithiodisulfide oxidoreductase DSBA Gamma interferon inducible lysosomal thiol reductase GILT Gamma interferon inducible lysosomal thiol reductase GILT Glutathione peroxidase GPX Glutathione peroxidase GPX Phospholipid hydroperoxide glutathione peroxidase PHGPX Glutathione peroxidase GPX Glutathione peroxidase GPX Glutathione peroxidase GPX Iodothyronine deiodinase DIO Iodothyronine deiodinase DIO Iodothyronine deiodinase DIO Iodothyronine deiodinase DIO Iron-sulfur oxidoreductase GLPC Membrane selenoprotein MSP Methylated-DNA-[protein]-cysteine-Smethyltransferase MGMT Peptide methionine-s-sulfoxide reductase MSRA Peptide methionine-s-sulfoxide reductase MSRA Protein disulfide isomerase PDI Protein disulfide isomerase PDI Protein disulfide isomerase PDI Selenoprotein 15 SEL Selenoprotein H SELH Selenoprotein H SELH Selenoprotein M SELM Selenoprotein M SELM Selenoprotein M SELM Selenoprotein M SELM Selenoprotein M SELM Selenoprotein M SELM Selenoprotein O SELO Selenoprotein O SELO Selenoprotein O SELO Selenoprotein O SELO Selenoprotein T SELT Selenoprotein T SELT
34 Selenoprotein U SELU Selenoprotein U SELU Selenoprotein U SELU Selenoprotein W SELW Thiol protein THIO Thiol protein THIO Thiol protein THIO Thiol:disulfide interchange protein TDIP Thioredoxin reductase TXR
35 Supplementary Table 15 Genes encoding proteins involved in the metabolism or synthesis of vitamins. Vitamin Name PID Core/Variable Pro-Vitamin A IPP/DMAPP Isomerase core GGPP synthase core core Phytoene synthase core Phytoene desaturase nd core Carotene desaturase core -cyclase core core B 1 Synthesis Dinucleotide-utilizing enzyme involved in molybdopterin and thiamine biosynthesis variable core core Thiamine monophosphate synthase core core variable Thiamine pyrophosphokinase core nd Doxyxylulose-5-phosphate synthase nd ABC-Type thiamine transporter core nd core B 1 Related (thiamine) Thiamine monophosphate synthase core Thiamine triphosphatase core nd Thiamine pyrophosphate enzyme-like TPP binding protein core variable Acetohydroxyacid synthase core nd nd variable core Plastid transketolase core 35
36 Transketolase core core core B 2 (Riboflavin) GTP cyclohydrolase II core 3,4-dihydroxy-2-butanone 4-phosphate synthase nd Bifunctional 2,5-diamino-6-ribosylamino-4 (3H)-pyrimidinone 5'- phosphate deaminase/reductase nd core 6,7-dimethyl-8-ribityllumazine synthase core Riboflavin synthase core Riboflavin kinase core core nd nd FAD synthetase core B 3 (Niacin) Aspartate oxidase core Quinolinate synthase Quinolinate phosphoribosyltransferase core B 5 (Pantothenic acid) Bifunctional ketopantoate hydroxymethyltransferase/panthothenate synthase variable Ketopantoate reductase nd B 6 Salvage Pathway (pyridoxal related compounds) Pyridoxal biosynthesis protein nd core core Probable glutamine aminotransferase core variable Pyrodoxal kinase core core variable Pyridoxal reductase core B 7 Metabolism (Biotin) 8-amino-7-oxononanoate synthase core core variable 36
37 Bifunctional detiohiotin synthetase/7,8-diamino-pelargonic acid aminotransferase core Biotin synthase variable DAPA core B 9 (folic Acid) GTP cyclohydrolase I core 5-aminoimidazole ribonucleotide carboxylase core 5-phosphoribosyl-1- pyrophosphate synthase/6- hydroxymethyldihydropterin pyrophosphokinase nd variable core 4-amino-4-deoxychorismate synthase core nd Dihydrofolate reductase/synthase core Dihydroneotperin (DHN) aldolase nd core Aminodeoxychorismate lyase core Dihydropteroate synthase variable core variable Folylpolyglutamate synthase nd core UDP-glucose-p-aminobenzoate glycosyltransferase core variable -glutamylhydrolase core nd B 12 Related (Cobalamin) S-adenosyl homocysteine hydrolase core core core Glycine hydroxymethyltransferase core core S-adenosylmethioine synthetase core core Homocystein S-methyltransferase core core Methylmalonyl-CoA mutase core Methylenetrtrahydrofolate reductase core core B12 Synthesis/Acquisition Uroporphyrin-III C/tetrapyrrole (Corrin/Porphyrin) methyltransferase core core 37
38 55801 variable Cobyrinic acid a,c-diamide synthase variable core Cobalamin adensyltransferase core nd Colalamin biosynthesis protein CobN and Mg=chelatases core core Cobalamin-5-phosphate synthase core Nicotinate-nucleotide-dimethylbenzimidazole core nd nd Cobalamin synthesis protein core nd core core core core core core core nd core core core nd nd variable core core variable core core core core variable core core Vitamin E Tocopherol Synthesis Homogentisate Phytyl transferase (VTE2) nd 38
39 55059 core nd Solanyl synthase Phytol kinase core core Tocopherol cyclase (VTE1) core core nd Tocopherol O-methyltransferase core core MPBQ/MSBQ methyltransferase core Phytyltransferase nd core nd variable C (Ascorbate) GTP-mannose pyrophosphorylase nd core GTP Mannose-3,5-epimerase/phosphmannose isomerase core nd L-galactose guanyltransferase L-galactose-1-P phosphatase core core L-galactose dehydrogenase core core L-galactono-1, 4-lactone dehydrogenase core nd D (calciferol) D3 24-hydoxylase variable 25-hydroxyvitamin D-1 alpha hydrolase variable Calcidiol 1- monooxygenase variable core core variable core core variable E (tocopherols) Homogentisate geranylgeranyl transferase nd core 39
40 56139 nd variable MPBQ/MSBQ methyltransferase core core Homogentisate solanyltransferase transferase core variable nd core Tocopherol cyclase core core nd Chorismate mutase core Hydroxyphenylpyruvate (HPP) dioxygenase core nd Tocopherol methyltransferase core K (Phylloquinone) Isochorismate synthase core PHYLLO core 1,4-dihydroxy-2-naphtoate phytyl transferase O-succinylbenzoate CoA ligase 1,4-dihydroxy-2-naphthoate CoA synthase 1,4-dihydroxy-2-naphthoate CoA thioesterase 1,4-dihydroxy-2-naphthoate phytyptransferase Demethylphylloquinone methyltransferase core core Vitamin K epoxide reductase core 40
tgg agg Supplementary Figure S1.
 ttaggatattcggtgaggtgatatgtctctgtttggaaatgtctccgccattaactcaag tggaaagtgtatagtaatgaatctttcaagcacacagatcacttcaaaagactgtttcaa catcacctcaggacaaaaagatgtactctcatttggatgctgtgatgccatgggtcacag attgcaattcccaagtgcccgttcttttacaccaaaatcaaagaagaatatctccccttt
ttaggatattcggtgaggtgatatgtctctgtttggaaatgtctccgccattaactcaag tggaaagtgtatagtaatgaatctttcaagcacacagatcacttcaaaagactgtttcaa catcacctcaggacaaaaagatgtactctcatttggatgctgtgatgccatgggtcacag attgcaattcccaagtgcccgttcttttacaccaaaatcaaagaagaatatctccccttt
Supporting information
 Supporting information Figure S1. Carotenoid biosynthesis pathway in papaya fruit which is adopted from Blas et al. (2010) (reference 4) and Nisar et al. (2015) (reference 5). Carotenoids are synthesized
Supporting information Figure S1. Carotenoid biosynthesis pathway in papaya fruit which is adopted from Blas et al. (2010) (reference 4) and Nisar et al. (2015) (reference 5). Carotenoids are synthesized
Supplementary Table S1. Genes that are upregulated during iron toxicity. Foldchange
 Supplementary Table S1. Genes that are upregulated during iron toxicity. Locus tag Spy Number a Foldchange b Annotation c Iron efflux pmta SpyM3_1093 17.6 Iron efflux pump Arginine and serine catabolism
Supplementary Table S1. Genes that are upregulated during iron toxicity. Locus tag Spy Number a Foldchange b Annotation c Iron efflux pmta SpyM3_1093 17.6 Iron efflux pump Arginine and serine catabolism
Experimental Identification and Computational Characterization of a Novel. Extracellular Metalloproteinase Produced by Clostridium sordellii
 Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 207 Supplementary Information Experimental Identification and Computational Characterization of
Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 207 Supplementary Information Experimental Identification and Computational Characterization of
Supporting Information for
 Supporting Information for Analysis of Sogatella furcifera proteome that interact with P10 protein of Southern rice black-streaked dwarf virus Win Than*, Faliang Qin*, Wenwen Liu, Xifeng Wang ** State
Supporting Information for Analysis of Sogatella furcifera proteome that interact with P10 protein of Southern rice black-streaked dwarf virus Win Than*, Faliang Qin*, Wenwen Liu, Xifeng Wang ** State
Functional Genomics & Proteomics
 Functional Genomics & Proteomics Genome Sequences TCACAATTTAGACATCTAGTCTTCCACTTAAGCATATTTAGATTGTTTCCAGTTTTCAGCTTTTATGACTAAATCTTCTAAAATTGTTTTTCCCTAAATGTATATTTTAATTTGTCTCAGGAGTAGAATTTCTGAGTCATAAAGCGGT CATATGTATAAATTTTAGGTGCCTCATAGCTCTTCAAATAGTCATCCCATTTTATACATCCAGGCAATATATGAGAGTTCTTGGTGCTCCACATCTTAGCTAGGATTTGATGTCAACCAGTCTCTTTAATTTAGATATTCTAGTACAT
Functional Genomics & Proteomics Genome Sequences TCACAATTTAGACATCTAGTCTTCCACTTAAGCATATTTAGATTGTTTCCAGTTTTCAGCTTTTATGACTAAATCTTCTAAAATTGTTTTTCCCTAAATGTATATTTTAATTTGTCTCAGGAGTAGAATTTCTGAGTCATAAAGCGGT CATATGTATAAATTTTAGGTGCCTCATAGCTCTTCAAATAGTCATCCCATTTTATACATCCAGGCAATATATGAGAGTTCTTGGTGCTCCACATCTTAGCTAGGATTTGATGTCAACCAGTCTCTTTAATTTAGATATTCTAGTACAT
Methods S1. Sequences relevant to the constructed strains, Related to Figures 1-6.
 Methods S1. Sequences relevant to the constructed strains, Related to Figures 1-6. A. Promoter Sequences Gal4 binding sites are highlighted in the color referenced in Figure 1A when possible. Site 1: red,
Methods S1. Sequences relevant to the constructed strains, Related to Figures 1-6. A. Promoter Sequences Gal4 binding sites are highlighted in the color referenced in Figure 1A when possible. Site 1: red,
Supplementary information: Biocatalysis on the surface of Escherichia coli: melanin pigmentation of the cell. exterior
 Supplementary information: Biocatalysis on the surface of Escherichia coli: melanin pigmentation of the cell exterior Martin Gustavsson, David Hörnström, Susanna Lundh, Jaroslav Belotserkovsky, Gen Larsson
Supplementary information: Biocatalysis on the surface of Escherichia coli: melanin pigmentation of the cell exterior Martin Gustavsson, David Hörnström, Susanna Lundh, Jaroslav Belotserkovsky, Gen Larsson
Eukaryotic Comparative Genomics
 Eukaryotic Comparative Genomics Detecting Conserved Sequences Charles Darwin Motoo Kimura Evolution of Neutral DNA A A T C TA AT T G CT G T GA T T C A GA G T A G CA G T GA AT A GT C T T T GA T GT T G T
Eukaryotic Comparative Genomics Detecting Conserved Sequences Charles Darwin Motoo Kimura Evolution of Neutral DNA A A T C TA AT T G CT G T GA T T C A GA G T A G CA G T GA AT A GT C T T T GA T GT T G T
MALE ADULT FIBROBLAST LINE (82-6hTERT)
 Double-stranded methylation patterns of a 104-bp L1 promoter in DNAs from male and female fibroblasts, male leukocytes and female lymphoblastoid cells using hairpin-bisulfite PCR. Fifteen L1 sequences
Double-stranded methylation patterns of a 104-bp L1 promoter in DNAs from male and female fibroblasts, male leukocytes and female lymphoblastoid cells using hairpin-bisulfite PCR. Fifteen L1 sequences
FETAL FIBROBLASTS, PASSAGE 10
 Double-stranded methylation patterns of a 104-bp L1 promoter in DNAs from fetal fibroblast passages 10, 14, 17, and 22 using barcoded hairpinbisulfite PCR. Fifteen L1 sequences were analyzed for passages
Double-stranded methylation patterns of a 104-bp L1 promoter in DNAs from fetal fibroblast passages 10, 14, 17, and 22 using barcoded hairpinbisulfite PCR. Fifteen L1 sequences were analyzed for passages
Plasmid Name: pmm290. Aliases: none known. Length: bp. Constructed by: Mike Moser/Cristina Swanson. Last updated: 17 August 2009
 Plasmid Name: pmm290 Aliases: none known Length: 11707 bp Constructed by: Mike Moser/Cristina Swanson Last updated: 17 August 2009 Description and application: This is a mammalian expression vector for
Plasmid Name: pmm290 Aliases: none known Length: 11707 bp Constructed by: Mike Moser/Cristina Swanson Last updated: 17 August 2009 Description and application: This is a mammalian expression vector for
Bounds on non-surjective cellular automata
 Bounds on non-surjective cellular automata Jarkko Kari Pascal Vanier Thomas Zeume University of Turku LIF Marseille Universität Hannover 27 august 2009 J. Kari, P. Vanier, T. Zeume (UTU) Bounds on non-surjective
Bounds on non-surjective cellular automata Jarkko Kari Pascal Vanier Thomas Zeume University of Turku LIF Marseille Universität Hannover 27 august 2009 J. Kari, P. Vanier, T. Zeume (UTU) Bounds on non-surjective
State of the Union... Functional Genomics Research Stream. Molecular Biology. Genomics. Computational Biology
 Functional Genomics Research Stream State of the Union... Research Meeting: February 16, 2010 Functional Genomics & Research Report III Concepts Genomics Molecular Biology Computational Biology Genome
Functional Genomics Research Stream State of the Union... Research Meeting: February 16, 2010 Functional Genomics & Research Report III Concepts Genomics Molecular Biology Computational Biology Genome
Efficiency change over time
 Efficiency change over time Heikki Tikanmäki Optimointiopin seminaari 14.11.2007 Contents Introduction (11.1) Window analysis (11.2) Example, application, analysis Malmquist index (11.3) Dealing with panel
Efficiency change over time Heikki Tikanmäki Optimointiopin seminaari 14.11.2007 Contents Introduction (11.1) Window analysis (11.2) Example, application, analysis Malmquist index (11.3) Dealing with panel
7.4 Variability management
 7.4 Variability management time... space software product-line should support variability in space (different products) support variability in time (maintenance, evolution) 1 Product variation Product
7.4 Variability management time... space software product-line should support variability in space (different products) support variability in time (maintenance, evolution) 1 Product variation Product
Table S2. S. thermophilus CRISPR spacer sequence analysis.
 Table S2. S. thermophilus CRISPR analysis. 001_1_01 TGTTTGACAGCAAATCAAGATTCGAATTGT 30 AF115103 Streptococcus thermophilus bacteriophage Sfi21, compl 40 93 28 2 0 3 30 38304 38331 1 001_1_02 AATGACGAGGAGCTATTGGCACAACTTACA
Table S2. S. thermophilus CRISPR analysis. 001_1_01 TGTTTGACAGCAAATCAAGATTCGAATTGT 30 AF115103 Streptococcus thermophilus bacteriophage Sfi21, compl 40 93 28 2 0 3 30 38304 38331 1 001_1_02 AATGACGAGGAGCTATTGGCACAACTTACA
Alternative DEA Models
 Mat-2.4142 Alternative DEA Models 19.9.2007 Table of Contents Banker-Charnes-Cooper Model Additive Model Example Data Home assignment BCC Model (Banker-Charnes-Cooper) production frontiers spanned by convex
Mat-2.4142 Alternative DEA Models 19.9.2007 Table of Contents Banker-Charnes-Cooper Model Additive Model Example Data Home assignment BCC Model (Banker-Charnes-Cooper) production frontiers spanned by convex
I. Principles of Pointer Year Analysis
 I. Principles of Pointer Year Analysis Fig 1. Maximum (red) and minimum (blue) pointer years. 1 Fig 2. Principle of pointer year calculation. Fig 3. Skeleton plot graph created by Kinsys/Kigraph programme.
I. Principles of Pointer Year Analysis Fig 1. Maximum (red) and minimum (blue) pointer years. 1 Fig 2. Principle of pointer year calculation. Fig 3. Skeleton plot graph created by Kinsys/Kigraph programme.
Results on the new polydrug use questions in the Finnish TDI data
 Results on the new polydrug use questions in the Finnish TDI data Multi-drug use, polydrug use and problematic polydrug use Martta Forsell, Finnish Focal Point 28/09/2015 Martta Forsell 1 28/09/2015 Esityksen
Results on the new polydrug use questions in the Finnish TDI data Multi-drug use, polydrug use and problematic polydrug use Martta Forsell, Finnish Focal Point 28/09/2015 Martta Forsell 1 28/09/2015 Esityksen
rapid evolution in copy number, location and sequence, with diverse turnover mechanisms.
 Repetitive DNA and its evolution: genomic strategies to understand the processes and exploit biodiversity rapid evolution in copy number, location and sequence, with diverse turnover mechanisms. often
Repetitive DNA and its evolution: genomic strategies to understand the processes and exploit biodiversity rapid evolution in copy number, location and sequence, with diverse turnover mechanisms. often
Capacity Utilization
 Capacity Utilization Tim Schöneberg 28th November Agenda Introduction Fixed and variable input ressources Technical capacity utilization Price based capacity utilization measure Long run and short run
Capacity Utilization Tim Schöneberg 28th November Agenda Introduction Fixed and variable input ressources Technical capacity utilization Price based capacity utilization measure Long run and short run
Genome 373: Genomic Informatics. Professors Elhanan Borenstein and Jay Shendure
 Genome 373: Genomic Informatics Professors Elhanan Borenstein and Jay Shendure Genome 373 This course is intended to introduce students to the breadth of problems and methods in computational analysis
Genome 373: Genomic Informatics Professors Elhanan Borenstein and Jay Shendure Genome 373 This course is intended to introduce students to the breadth of problems and methods in computational analysis
Other approaches to restrict multipliers
 Other approaches to restrict multipliers Heikki Tikanmäki Optimointiopin seminaari 10.10.2007 Contents Short revision (6.2) Another Assurance Region Model (6.3) Cone-Ratio Method (6.4) An Application of
Other approaches to restrict multipliers Heikki Tikanmäki Optimointiopin seminaari 10.10.2007 Contents Short revision (6.2) Another Assurance Region Model (6.3) Cone-Ratio Method (6.4) An Application of
Chapter 7. Motif finding (week 11) Chapter 8. Sequence binning (week 11)
 Course organization Introduction ( Week 1) Part I: Algorithms for Sequence Analysis (Week 1-11) Chapter 1-3, Models and theories» Probability theory and Statistics (Week 2)» Algorithm complexity analysis
Course organization Introduction ( Week 1) Part I: Algorithms for Sequence Analysis (Week 1-11) Chapter 1-3, Models and theories» Probability theory and Statistics (Week 2)» Algorithm complexity analysis
Metsälamminkankaan tuulivoimapuiston osayleiskaava
 VAALAN KUNTA TUULISAIMAA OY Metsälamminkankaan tuulivoimapuiston osayleiskaava Liite 3. Varjostusmallinnus FCG SUUNNITTELU JA TEKNIIKKA OY 12.5.2015 P25370 SHADOW - Main Result Assumptions for shadow calculations
VAALAN KUNTA TUULISAIMAA OY Metsälamminkankaan tuulivoimapuiston osayleiskaava Liite 3. Varjostusmallinnus FCG SUUNNITTELU JA TEKNIIKKA OY 12.5.2015 P25370 SHADOW - Main Result Assumptions for shadow calculations
16. Allocation Models
 16. Allocation Models Juha Saloheimo 17.1.27 S steemianalsin Optimointiopin seminaari - Sks 27 Content Introduction Overall Efficienc with common prices and costs Cost Efficienc S steemianalsin Revenue
16. Allocation Models Juha Saloheimo 17.1.27 S steemianalsin Optimointiopin seminaari - Sks 27 Content Introduction Overall Efficienc with common prices and costs Cost Efficienc S steemianalsin Revenue
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
Returns to Scale II. S ysteemianalyysin. Laboratorio. Esitelmä 8 Timo Salminen. Teknillinen korkeakoulu
 Returns to Scale II Contents Most Productive Scale Size Further Considerations Relaxation of the Convexity Condition Useful Reminder Theorem 5.5 A DMU found to be efficient with a CCR model will also be
Returns to Scale II Contents Most Productive Scale Size Further Considerations Relaxation of the Convexity Condition Useful Reminder Theorem 5.5 A DMU found to be efficient with a CCR model will also be
Basset: Learning the regulatory code of the accessible genome with deep convolutional neural networks. David R. Kelley
 Basset: Learning the regulatory code of the accessible genome with deep convolutional neural networks David R. Kelley DNA codes for complex life. How? Kundaje et al. Integrative analysis of 111 reference
Basset: Learning the regulatory code of the accessible genome with deep convolutional neural networks David R. Kelley DNA codes for complex life. How? Kundaje et al. Integrative analysis of 111 reference
Strain or plasmid Description a Reference or source b. 168 trpc2 Laboratory. 1A780 trpc2 sigb::spc BGSC
 TABLE S1 Bacterial strains and plasmids Strain or plasmid Description a Reference or source b B. subtilis 168 trpc2 Laboratory stock 1A780 trpc2 sigb::spc BGSC BM1010 trpc2 htpx::pgs1582; Em r BM1302 trpc2
TABLE S1 Bacterial strains and plasmids Strain or plasmid Description a Reference or source b B. subtilis 168 trpc2 Laboratory stock 1A780 trpc2 sigb::spc BGSC BM1010 trpc2 htpx::pgs1582; Em r BM1302 trpc2
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
LX 70. Ominaisuuksien mittaustulokset 1-kerroksinen 2-kerroksinen. Fyysiset ominaisuudet, nimellisarvot. Kalvon ominaisuudet
 LX 70 % Läpäisy 36 32 % Absorptio 30 40 % Heijastus 34 28 % Läpäisy 72 65 % Heijastus ulkopuoli 9 16 % Heijastus sisäpuoli 9 13 Emissiivisyys.77.77 Auringonsuojakerroin.54.58 Auringonsäteilyn lämmönsiirtokerroin.47.50
LX 70 % Läpäisy 36 32 % Absorptio 30 40 % Heijastus 34 28 % Läpäisy 72 65 % Heijastus ulkopuoli 9 16 % Heijastus sisäpuoli 9 13 Emissiivisyys.77.77 Auringonsuojakerroin.54.58 Auringonsäteilyn lämmönsiirtokerroin.47.50
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
Gap-filling methods for CH 4 data
 Gap-filling methods for CH 4 data Sigrid Dengel University of Helsinki Outline - Ecosystems known for CH 4 emissions; - Why is gap-filling of CH 4 data not as easy and straight forward as CO 2 ; - Gap-filling
Gap-filling methods for CH 4 data Sigrid Dengel University of Helsinki Outline - Ecosystems known for CH 4 emissions; - Why is gap-filling of CH 4 data not as easy and straight forward as CO 2 ; - Gap-filling
A DEA Game II. Juha Saloheimo S ysteemianalyysin. Laboratorio. Teknillinen korkeakoulu
 A DEA Game II Juha Salohemo 12.12.2007 Content Recap of the Example The Shapley Value Margnal Contrbuton, Ordered Coaltons, Soluton to the Example DEA Mn Game Summary Home Assgnment Recap of the Example
A DEA Game II Juha Salohemo 12.12.2007 Content Recap of the Example The Shapley Value Margnal Contrbuton, Ordered Coaltons, Soluton to the Example DEA Mn Game Summary Home Assgnment Recap of the Example
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.9.269
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.9.269
Rgg-Shp regulators are important for pneumococcal colonization and invasion. through their effect on mannose utilization and capsule synthesis
 Supplementary information Rgg-Shp regulators are important for pneumococcal colonization and invasion through their effect on mannose utilization and capsule synthesis Xiangyun Zhi 1, Iman Tajer Abdullah
Supplementary information Rgg-Shp regulators are important for pneumococcal colonization and invasion through their effect on mannose utilization and capsule synthesis Xiangyun Zhi 1, Iman Tajer Abdullah
( ( OX2 Perkkiö. Rakennuskanta. Varjostus. 9 x N131 x HH145
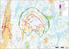 OX2 9 x N131 x HH145 Rakennuskanta Asuinrakennus Lomarakennus Liike- tai julkinen rakennus Teollinen rakennus Kirkko tai kirkollinen rak. Muu rakennus Allas Varjostus 1 h/a 8 h/a 20 h/a 0 0,5 1 1,5 2 km
OX2 9 x N131 x HH145 Rakennuskanta Asuinrakennus Lomarakennus Liike- tai julkinen rakennus Teollinen rakennus Kirkko tai kirkollinen rak. Muu rakennus Allas Varjostus 1 h/a 8 h/a 20 h/a 0 0,5 1 1,5 2 km
National Building Code of Finland, Part D1, Building Water Supply and Sewerage Systems, Regulations and guidelines 2007
 National Building Code of Finland, Part D1, Building Water Supply and Sewerage Systems, Regulations and guidelines 2007 Chapter 2.4 Jukka Räisä 1 WATER PIPES PLACEMENT 2.4.1 Regulation Water pipe and its
National Building Code of Finland, Part D1, Building Water Supply and Sewerage Systems, Regulations and guidelines 2007 Chapter 2.4 Jukka Räisä 1 WATER PIPES PLACEMENT 2.4.1 Regulation Water pipe and its
Regulatory Sequence Analysis. Discovering phylogenetic footprints in bacterial promoters
 Regulatory Sequence Analysis Discovering phylogenetic footprints in bacterial promoters DNA-protein binding interface Example of DNA-protein interface. LexA homodimer on the reca promoter. Note: this is
Regulatory Sequence Analysis Discovering phylogenetic footprints in bacterial promoters DNA-protein binding interface Example of DNA-protein interface. LexA homodimer on the reca promoter. Note: this is
Tynnyrivaara, OX2 Tuulivoimahanke. ( Layout 9 x N131 x HH145. Rakennukset Asuinrakennus Lomarakennus 9 x N131 x HH145 Varjostus 1 h/a 8 h/a 20 h/a
 , Tuulivoimahanke Layout 9 x N131 x HH145 Rakennukset Asuinrakennus Lomarakennus 9 x N131 x HH145 Varjostus 1 h/a 8 h/a 20 h/a 0 0,5 1 1,5 km 2 SHADOW - Main Result Assumptions for shadow calculations
, Tuulivoimahanke Layout 9 x N131 x HH145 Rakennukset Asuinrakennus Lomarakennus 9 x N131 x HH145 Varjostus 1 h/a 8 h/a 20 h/a 0 0,5 1 1,5 km 2 SHADOW - Main Result Assumptions for shadow calculations
Telecommunication Software
 Telecommunication Software Final exam 21.11.2006 COMPUTER ENGINEERING LABORATORY 521265A Vastaukset englanniksi tai suomeksi. / Answers in English or in Finnish. 1. (a) Määrittele sovellusviesti, PersonnelRecord,
Telecommunication Software Final exam 21.11.2006 COMPUTER ENGINEERING LABORATORY 521265A Vastaukset englanniksi tai suomeksi. / Answers in English or in Finnish. 1. (a) Määrittele sovellusviesti, PersonnelRecord,
Valmiustaitoja biokemisteille
 Valmiustaitoja biokemisteille - Power Point -ohjelman käyttö - seminaariesitelmän laatiminen ja esittäminen Tuomo Glumoff 1. Power Point -ohjelman käyttö - teksti - kuvat - tausta - valmiit pohjat * löytyy
Valmiustaitoja biokemisteille - Power Point -ohjelman käyttö - seminaariesitelmän laatiminen ja esittäminen Tuomo Glumoff 1. Power Point -ohjelman käyttö - teksti - kuvat - tausta - valmiit pohjat * löytyy
7. Product-line architectures
 7. Product-line architectures 7.1 Introduction 7.2 Product-line basics 7.3 Layered style for product-lines 7.4 Variability management 7.5 Benefits and problems with product-lines 1 Short history of software
7. Product-line architectures 7.1 Introduction 7.2 Product-line basics 7.3 Layered style for product-lines 7.4 Variability management 7.5 Benefits and problems with product-lines 1 Short history of software
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
Bioinformatics. Sequence Analysis: Part III. Pattern Searching and Gene Finding. Fran Lewitter, Ph.D. Head, Biocomputing Whitehead Institute
 Bioinformatics Sequence Analysis: Part III. Pattern Searching and Gene Finding Fran Lewitter, Ph.D. Head, Biocomputing Whitehead Institute Course Syllabus Jan 7 Jan 14 Jan 21 Jan 28 Feb 4 Feb 11 Feb 18
Bioinformatics Sequence Analysis: Part III. Pattern Searching and Gene Finding Fran Lewitter, Ph.D. Head, Biocomputing Whitehead Institute Course Syllabus Jan 7 Jan 14 Jan 21 Jan 28 Feb 4 Feb 11 Feb 18
Lampiran 1 Komposisi media pertumbuhan yang digunakan A. Media nutrien agar (NA) - Agar 1.5% - Nutrient broth 0.8%
 LAMPIRAN 8 9 Lampiran 1 Komposisi media pertumbuhan yang digunakan A. Media nutrien agar (NA) - Agar 1.5% - Nutrient broth 0.8% B. Media King s B agar - Agar 1.5% - Pepton 2% - K 2 HPO 4 0.15% - MgSO 4.4H
LAMPIRAN 8 9 Lampiran 1 Komposisi media pertumbuhan yang digunakan A. Media nutrien agar (NA) - Agar 1.5% - Nutrient broth 0.8% B. Media King s B agar - Agar 1.5% - Pepton 2% - K 2 HPO 4 0.15% - MgSO 4.4H
Title: Enhancement of protein production via the strong DIT1 terminator and two RNA-binding proteins in Saccharomyces cerevisiae
 Title: Enhancement of protein production via the strong DIT1 terminator and two RNA-binding proteins in Saccharomyces cerevisiae Authors: Yoichiro Ito, Takao Kitagawa, Mamoru Yamanishi, Satoshi Katahira,
Title: Enhancement of protein production via the strong DIT1 terminator and two RNA-binding proteins in Saccharomyces cerevisiae Authors: Yoichiro Ito, Takao Kitagawa, Mamoru Yamanishi, Satoshi Katahira,
Underpinning Phage Arbitrium Communication Systems
 Molecular Cell, Volume 74 Supplemental Information Deciphering the Molecular Mechanism Underpinning Phage Arbitrium Communication Systems Francisca Gallego del Sol, José R. Penadés, and Alberto Marina
Molecular Cell, Volume 74 Supplemental Information Deciphering the Molecular Mechanism Underpinning Phage Arbitrium Communication Systems Francisca Gallego del Sol, José R. Penadés, and Alberto Marina
TM ETRS-TM35FIN-ETRS89 WTG
 VE1 SHADOW - Main Result Calculation: 8 x Nordex N131 x HH145m Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please
VE1 SHADOW - Main Result Calculation: 8 x Nordex N131 x HH145m Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please
KMTK lentoestetyöpaja - Osa 2
 KMTK lentoestetyöpaja - Osa 2 Veijo Pätynen 18.10.2016 Pasila YHTEISTYÖSSÄ: Ilmailun paikkatiedon hallintamalli Ilmailun paikkatiedon hallintamalli (v0.9 4.3.2016) 4.4 Maanmittauslaitoksen rooli ja vastuut...
KMTK lentoestetyöpaja - Osa 2 Veijo Pätynen 18.10.2016 Pasila YHTEISTYÖSSÄ: Ilmailun paikkatiedon hallintamalli Ilmailun paikkatiedon hallintamalli (v0.9 4.3.2016) 4.4 Maanmittauslaitoksen rooli ja vastuut...
WindPRO version joulu 2012 Printed/Page :42 / 1. SHADOW - Main Result
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table 13.6.2013 19:42 / 1 Minimum
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table 13.6.2013 19:42 / 1 Minimum
 Digitally signed by Hans Vadbäck DN: cn=hans Vadbäck, o, ou=fcg Suunnittelu ja Tekniikka Oy, email=hans.vadback@fcg.fi, c=fi Date: 2016.12.20 15:45:35 +02'00' Jakob Kjellman Digitally signed by Jakob Kjellman
Digitally signed by Hans Vadbäck DN: cn=hans Vadbäck, o, ou=fcg Suunnittelu ja Tekniikka Oy, email=hans.vadback@fcg.fi, c=fi Date: 2016.12.20 15:45:35 +02'00' Jakob Kjellman Digitally signed by Jakob Kjellman
ReFuel 70 % Emission Reduction Using Renewable High Cetane Number Paraffinic Diesel Fuel. Kalle Lehto, Aalto-yliopisto 5.5.
 ReFuel 70 % Emission Reduction Using Renewable High Cetane Number Paraffinic Diesel Fuel Kalle Lehto, Aalto-yliopisto 5.5.2011 Otaniemi ReFuel a three year research project (2009-2011) goal utilize the
ReFuel 70 % Emission Reduction Using Renewable High Cetane Number Paraffinic Diesel Fuel Kalle Lehto, Aalto-yliopisto 5.5.2011 Otaniemi ReFuel a three year research project (2009-2011) goal utilize the
( ,5 1 1,5 2 km
 Tuulivoimala Rakennukset Asuinrakennus Liikerak. tai Julkinen rak. Lomarakennus Teollinen rakennus Kirkollinen rakennus Varjostus "real case" h/a 1 h/a 8 h/a 20 h/a 4 5 3 1 2 6 7 8 9 10 0 0,5 1 1,5 2 km
Tuulivoimala Rakennukset Asuinrakennus Liikerak. tai Julkinen rak. Lomarakennus Teollinen rakennus Kirkollinen rakennus Varjostus "real case" h/a 1 h/a 8 h/a 20 h/a 4 5 3 1 2 6 7 8 9 10 0 0,5 1 1,5 2 km
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table 22.12.2014 11:33 / 1 Minimum
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table 22.12.2014 11:33 / 1 Minimum
,0 Yes ,0 120, ,8
 SHADOW - Main Result Calculation: Alue 2 ( x 9 x HH120) TuuliSaimaa kaavaluonnos Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered
SHADOW - Main Result Calculation: Alue 2 ( x 9 x HH120) TuuliSaimaa kaavaluonnos Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered
Predicting evolutionarily conserved regions (ECRs) in the Xenopus tropicalis genome using a MultiPipMaker-based bioinformatic strategy
 Predicting evolutionarily conserved regions (ECRs) in the Xenopus tropicalis genome using a MultiPipMaker-based bioinformatic strategy OVERVIEW- Method updated and written by Sarah Louie (6/19/08) Initially
Predicting evolutionarily conserved regions (ECRs) in the Xenopus tropicalis genome using a MultiPipMaker-based bioinformatic strategy OVERVIEW- Method updated and written by Sarah Louie (6/19/08) Initially
T Statistical Natural Language Processing Answers 6 Collocations Version 1.0
 T-61.5020 Statistical Natural Language Processing Answers 6 Collocations Version 1.0 1. Let s start by calculating the results for pair valkoinen, talo manually: Frequency: Bigrams valkoinen, talo occurred
T-61.5020 Statistical Natural Language Processing Answers 6 Collocations Version 1.0 1. Let s start by calculating the results for pair valkoinen, talo manually: Frequency: Bigrams valkoinen, talo occurred
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
Capacity utilization
 Mat-2.4142 Seminar on optimization Capacity utilization 12.12.2007 Contents Summary of chapter 14 Related DEA-solver models Illustrative examples Measure of technical capacity utilization Price-based measure
Mat-2.4142 Seminar on optimization Capacity utilization 12.12.2007 Contents Summary of chapter 14 Related DEA-solver models Illustrative examples Measure of technical capacity utilization Price-based measure
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Calculation: N117 x 9 x HH141 Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG
SHADOW - Main Result Calculation: N117 x 9 x HH141 Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG
KONEISTUSKOKOONPANON TEKEMINEN NX10-YMPÄRISTÖSSÄ
 KONEISTUSKOKOONPANON TEKEMINEN NX10-YMPÄRISTÖSSÄ https://community.plm.automation.siemens.com/t5/tech-tips- Knowledge-Base-NX/How-to-simulate-any-G-code-file-in-NX- CAM/ta-p/3340 Koneistusympäristön määrittely
KONEISTUSKOKOONPANON TEKEMINEN NX10-YMPÄRISTÖSSÄ https://community.plm.automation.siemens.com/t5/tech-tips- Knowledge-Base-NX/How-to-simulate-any-G-code-file-in-NX- CAM/ta-p/3340 Koneistusympäristön määrittely
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table 5.11.2013 16:44 / 1 Minimum
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table 5.11.2013 16:44 / 1 Minimum
VIIKKI BIOCENTER University of Helsinki
 VIIKKI BIOCENTER University of Helsinki Biologian DNA koodi ja sen selvittäminen Petri Auvinen DNA Sequencing and Genomics Laboratory Institute of Biotechnology Kuinka solut kehittyivät? Kolmenlaisia soluja
VIIKKI BIOCENTER University of Helsinki Biologian DNA koodi ja sen selvittäminen Petri Auvinen DNA Sequencing and Genomics Laboratory Institute of Biotechnology Kuinka solut kehittyivät? Kolmenlaisia soluja
VIIKKI BIOCENTER University of Helsinki
 VIIKKI BIOCENTER University of Helsinki Mitä uudet DNA sekvensointimenetelmät voivat paljastaa luonnonjärjestelmästä? Petri Auvinen DNA Sequencing and Genomics Laboratory Institute of Biotechnology Petri
VIIKKI BIOCENTER University of Helsinki Mitä uudet DNA sekvensointimenetelmät voivat paljastaa luonnonjärjestelmästä? Petri Auvinen DNA Sequencing and Genomics Laboratory Institute of Biotechnology Petri
ABB Oy Domestic Sales 20.10.2015 Harri Liukku Aurinkosähköjärjestelmät Kytkennät
 ABB Oy Domestic Sales 20.10.2015 Harri Liukku Aurinkosähköjärjestelmät Kytkennät ABB Group ABB on johtava sähkövoima- ja automaatioteknologiayhtymä. Potentiaalisina kasvualueina uusiutuva energia ja liikenteen
ABB Oy Domestic Sales 20.10.2015 Harri Liukku Aurinkosähköjärjestelmät Kytkennät ABB Group ABB on johtava sähkövoima- ja automaatioteknologiayhtymä. Potentiaalisina kasvualueina uusiutuva energia ja liikenteen
HARJOITUS- PAKETTI A
 Logistiikka A35A00310 Tuotantotalouden perusteet HARJOITUS- PAKETTI A (6 pistettä) TUTA 19 Luento 3.Ennustaminen County General 1 piste The number of heart surgeries performed at County General Hospital
Logistiikka A35A00310 Tuotantotalouden perusteet HARJOITUS- PAKETTI A (6 pistettä) TUTA 19 Luento 3.Ennustaminen County General 1 piste The number of heart surgeries performed at County General Hospital
Supplementary Table S1. Material list (a) Parameters Sal to Str
 Tooth wear as a means to quantify intra-specific variations in diet and chewing movements - Scientific Reports 2016, 6:3037 Ivan Calandra, Gaëlle Labonne, Ellen Schulz-Kornas, Thomas M. Kaiser & Sophie
Tooth wear as a means to quantify intra-specific variations in diet and chewing movements - Scientific Reports 2016, 6:3037 Ivan Calandra, Gaëlle Labonne, Ellen Schulz-Kornas, Thomas M. Kaiser & Sophie
MIKES, Julkaisu J3/2000 MASS COMPARISON M3. Comparison of 1 kg and 10 kg weights between MIKES and three FINAS accredited calibration laboratories
 MITTATEKNIIKAN KESKUS CENTRE FOR METROLOGY AND ACCREDITATION Julkaisu J3/2000 MASS COMPARISON M3 Comparison of 1 kg and 10 kg weights between MIKES and three FINAS accredited calibration laboratories Kari
MITTATEKNIIKAN KESKUS CENTRE FOR METROLOGY AND ACCREDITATION Julkaisu J3/2000 MASS COMPARISON M3 Comparison of 1 kg and 10 kg weights between MIKES and three FINAS accredited calibration laboratories Kari
WindPRO version joulu 2012 Printed/Page :47 / 1. SHADOW - Main Result
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
Research plan for masters thesis in forest sciences. The PELLETime 2009 Symposium Mervi Juntunen
 Modelling tree and stand characteristics and estimating biomass removals and harvesting costs of lodgepole pine (Pinus contorta) plantations in Iceland Research plan for masters thesis in forest sciences
Modelling tree and stand characteristics and estimating biomass removals and harvesting costs of lodgepole pine (Pinus contorta) plantations in Iceland Research plan for masters thesis in forest sciences
B3206 Microbial Genetics
 Prof. Fahd M. Nasr Lebanese university Faculty of sciences I Department of Natural Sciences fnasr@ul.edu.lb B3206 Microbial Genetics 1 Eukaryotic M. G. The yeast Saccharomyces cerevisiae as a genetic model
Prof. Fahd M. Nasr Lebanese university Faculty of sciences I Department of Natural Sciences fnasr@ul.edu.lb B3206 Microbial Genetics 1 Eukaryotic M. G. The yeast Saccharomyces cerevisiae as a genetic model
Use of spatial data in the new production environment and in a data warehouse
 Use of spatial data in the new production environment and in a data warehouse Nordic Forum for Geostatistics 2007 Session 3, GI infrastructure and use of spatial database Statistics Finland, Population
Use of spatial data in the new production environment and in a data warehouse Nordic Forum for Geostatistics 2007 Session 3, GI infrastructure and use of spatial database Statistics Finland, Population
The CCR Model and Production Correspondence
 The CCR Model and Production Correspondence Tim Schöneberg The 19th of September Agenda Introduction Definitions Production Possiblity Set CCR Model and the Dual Problem Input excesses and output shortfalls
The CCR Model and Production Correspondence Tim Schöneberg The 19th of September Agenda Introduction Definitions Production Possiblity Set CCR Model and the Dual Problem Input excesses and output shortfalls
Viral DNA as a model for coil to globule transition
 Viral DNA as a model for coil to globule transition Marina Rossi Lab. of complex fluids and molecular biophysics LITA (Segrate) UNIVERSITA DEGLI STUDI DI MILANO - PhD Workshop October 14 th, 2013 Temperature
Viral DNA as a model for coil to globule transition Marina Rossi Lab. of complex fluids and molecular biophysics LITA (Segrate) UNIVERSITA DEGLI STUDI DI MILANO - PhD Workshop October 14 th, 2013 Temperature
Rakennukset Varjostus "real case" h/a 0,5 1,5
 Tuulivoimala Rakennukset Asuinrakennus Liikerak. tai Julkinen rak. Lomarakennus Teollinen rakennus Kirkollinen rakennus Varjostus "real case" h/a 1 h/a 8 h/a 20 h/a 1 2 3 5 8 4 6 7 9 10 0 0,5 1 1,5 2 km
Tuulivoimala Rakennukset Asuinrakennus Liikerak. tai Julkinen rak. Lomarakennus Teollinen rakennus Kirkollinen rakennus Varjostus "real case" h/a 1 h/a 8 h/a 20 h/a 1 2 3 5 8 4 6 7 9 10 0 0,5 1 1,5 2 km
Enseigner l'évolution
 AATTTTCGTATCTGTTGGAGTTAGATAAGCCTACGCTTGATGGACCGTTGGGTGGCTTTCTAAGTGAGCTCGTGCCATCACAATTAATATAAGGAATTGTAGATGTTTCTTTCGTTATAGGTATTTCAAAAT TTATAAGAACCTACGCCCTCGTCTTTCTCCATTGGAACAGTTGCCGTTTTCGCAGTTCTTTTTGGTTCAGTCCTCATATCATGTGATTCCCCTGGCTCTCCTGATCTTTTTATACTTACTTTGAAATCGTCA
AATTTTCGTATCTGTTGGAGTTAGATAAGCCTACGCTTGATGGACCGTTGGGTGGCTTTCTAAGTGAGCTCGTGCCATCACAATTAATATAAGGAATTGTAGATGTTTCTTTCGTTATAGGTATTTCAAAAT TTATAAGAACCTACGCCCTCGTCTTTCTCCATTGGAACAGTTGCCGTTTTCGCAGTTCTTTTTGGTTCAGTCCTCATATCATGTGATTCCCCTGGCTCTCCTGATCTTTTTATACTTACTTTGAAATCGTCA
Tilausvahvistus. Anttolan Urheilijat HENNA-RIIKKA HAIKONEN KUMMANNIEMENTIE 5 B RAHULA. Anttolan Urheilijat
 7.80.4 Asiakasnumero: 3000359 KALLE MANNINEN KOVASTENLUODONTIE 46 51600 HAUKIVUORI Toimitusosoite: KUMMANNIEMENTIE 5 B 51720 RAHULA Viitteenne: Henna-Riikka Haikonen Viitteemme: Pyry Niemi +358400874498
7.80.4 Asiakasnumero: 3000359 KALLE MANNINEN KOVASTENLUODONTIE 46 51600 HAUKIVUORI Toimitusosoite: KUMMANNIEMENTIE 5 B 51720 RAHULA Viitteenne: Henna-Riikka Haikonen Viitteemme: Pyry Niemi +358400874498
Graph. COMPUTE x=rv.normal(0,0.04). COMPUTE y=rv.normal(0,0.04). execute.
 COMPUTE x=rv.ormal(0,0.04). COMPUTE y=rv.ormal(0,0.04). execute. compute hplib_man_r = hplib_man + x. compute arvokons_man_r = arvokons_man + y. GRAPH /SCATTERPLOT(BIVAR)=hplib_man_r WITH arvokons_man_r
COMPUTE x=rv.ormal(0,0.04). COMPUTE y=rv.ormal(0,0.04). execute. compute hplib_man_r = hplib_man + x. compute arvokons_man_r = arvokons_man + y. GRAPH /SCATTERPLOT(BIVAR)=hplib_man_r WITH arvokons_man_r
C++11 seminaari, kevät Johannes Koskinen
 C++11 seminaari, kevät 2012 Johannes Koskinen Sisältö Mikä onkaan ongelma? Standardidraftin luku 29: Atomiset tyypit Muistimalli Rinnakkaisuus On multicore systems, when a thread writes a value to memory,
C++11 seminaari, kevät 2012 Johannes Koskinen Sisältö Mikä onkaan ongelma? Standardidraftin luku 29: Atomiset tyypit Muistimalli Rinnakkaisuus On multicore systems, when a thread writes a value to memory,
ELEMET- MOCASTRO. Effect of grain size on A 3 temperatures in C-Mn and low alloyed steels - Gleeble tests and predictions. Period
 1 ELEMET- MOCASTRO Effect of grain size on A 3 temperatures in C-Mn and low alloyed steels - Gleeble tests and predictions Period 20.02-25.05.2012 Diaarinumero Rahoituspäätöksen numero 1114/31/2010 502/10
1 ELEMET- MOCASTRO Effect of grain size on A 3 temperatures in C-Mn and low alloyed steels - Gleeble tests and predictions Period 20.02-25.05.2012 Diaarinumero Rahoituspäätöksen numero 1114/31/2010 502/10
enzymatic families regulated in response to a short-term cytokinin treatment in root apices
 A B C D Supplemental tlfigure 1. 1Mtbli Metabolic processes, hormonal pathways and dlarge enzymatic families regulated in response to a short-term cytokinin treatment in root apices The 297 transcripts
A B C D Supplemental tlfigure 1. 1Mtbli Metabolic processes, hormonal pathways and dlarge enzymatic families regulated in response to a short-term cytokinin treatment in root apices The 297 transcripts
Paikkatiedon semanttinen mallinnus, integrointi ja julkaiseminen Case Suomalainen ajallinen paikkaontologia SAPO
 Paikkatiedon semanttinen mallinnus, integrointi ja julkaiseminen Case Suomalainen ajallinen paikkaontologia SAPO Tomi Kauppinen, Eero Hyvönen, Jari Väätäinen Semantic Computing Research Group (SeCo) http://www.seco.tkk.fi/
Paikkatiedon semanttinen mallinnus, integrointi ja julkaiseminen Case Suomalainen ajallinen paikkaontologia SAPO Tomi Kauppinen, Eero Hyvönen, Jari Väätäinen Semantic Computing Research Group (SeCo) http://www.seco.tkk.fi/
TIEKE Verkottaja Service Tools for electronic data interchange utilizers. Heikki Laaksamo
 TIEKE Verkottaja Service Tools for electronic data interchange utilizers Heikki Laaksamo TIEKE Finnish Information Society Development Centre (TIEKE Tietoyhteiskunnan kehittämiskeskus ry) TIEKE is a neutral,
TIEKE Verkottaja Service Tools for electronic data interchange utilizers Heikki Laaksamo TIEKE Finnish Information Society Development Centre (TIEKE Tietoyhteiskunnan kehittämiskeskus ry) TIEKE is a neutral,
FinFamily PostgreSQL installation ( ) FinFamily PostgreSQL
 FinFamily PostgreSQL 1 Sisällys / Contents FinFamily PostgreSQL... 1 1. Asenna PostgreSQL tietokanta / Install PostgreSQL database... 3 1.1. PostgreSQL tietokannasta / About the PostgreSQL database...
FinFamily PostgreSQL 1 Sisällys / Contents FinFamily PostgreSQL... 1 1. Asenna PostgreSQL tietokanta / Install PostgreSQL database... 3 1.1. PostgreSQL tietokannasta / About the PostgreSQL database...
The relationship between leisuretime physical activity and work stress with special reference to heart rate variability analyses
 The relationship between leisuretime physical activity and work stress with special reference to heart rate variability analyses Teisala Tiina, TtM, tohtorikoulutettava Jyväskylän yliopisto Terveystieteiden
The relationship between leisuretime physical activity and work stress with special reference to heart rate variability analyses Teisala Tiina, TtM, tohtorikoulutettava Jyväskylän yliopisto Terveystieteiden
Operatioanalyysi 2011, Harjoitus 2, viikko 38
 Operatioanalyysi 2011, Harjoitus 2, viikko 38 H2t1, Exercise 1.1. H2t2, Exercise 1.2. H2t3, Exercise 2.3. H2t4, Exercise 2.4. H2t5, Exercise 2.5. (Exercise 1.1.) 1 1.1. Model the following problem mathematically:
Operatioanalyysi 2011, Harjoitus 2, viikko 38 H2t1, Exercise 1.1. H2t2, Exercise 1.2. H2t3, Exercise 2.3. H2t4, Exercise 2.4. H2t5, Exercise 2.5. (Exercise 1.1.) 1 1.1. Model the following problem mathematically:
Sosiaalisessa mediassa syntyneen datan hyödyntäminen onnettomuuksissa ja muissa kriisitilanteissa
 Sosiaalisessa mediassa syntyneen datan hyödyntäminen onnettomuuksissa ja muissa kriisitilanteissa Aapo Immonen Jatko-opiskelija, Itä Suomen Yliopisto, Kansanterveystieteen Laitos aapoo@student.uef.fi Hannu
Sosiaalisessa mediassa syntyneen datan hyödyntäminen onnettomuuksissa ja muissa kriisitilanteissa Aapo Immonen Jatko-opiskelija, Itä Suomen Yliopisto, Kansanterveystieteen Laitos aapoo@student.uef.fi Hannu
Efficient regeneration system and Agrobacteriummediated transformation of Vetiver. PT 1
 Efficient regeneration system and Agrobacteriummediated transformation of Vetiver PT www.acolor.net 1 Outline: Part I: Why (Background) Part II: Some concrete work we have done Part III: Prospects to further
Efficient regeneration system and Agrobacteriummediated transformation of Vetiver PT www.acolor.net 1 Outline: Part I: Why (Background) Part II: Some concrete work we have done Part III: Prospects to further
1. SIT. The handler and dog stop with the dog sitting at heel. When the dog is sitting, the handler cues the dog to heel forward.
 START START SIT 1. SIT. The handler and dog stop with the dog sitting at heel. When the dog is sitting, the handler cues the dog to heel forward. This is a static exercise. SIT STAND 2. SIT STAND. The
START START SIT 1. SIT. The handler and dog stop with the dog sitting at heel. When the dog is sitting, the handler cues the dog to heel forward. This is a static exercise. SIT STAND 2. SIT STAND. The
Työmaadoitusvälineet suurjännitteelle
 Työmaadoitusvälineet suurjännitteelle Eurolaite Oy on vuonna 1 perustettu sähkötekniikan tuotteiden maahantuontiin, markkinointiin ja myyntiin erikoistunut asiantuntijayritys. Keskeisenä tavoitteena on
Työmaadoitusvälineet suurjännitteelle Eurolaite Oy on vuonna 1 perustettu sähkötekniikan tuotteiden maahantuontiin, markkinointiin ja myyntiin erikoistunut asiantuntijayritys. Keskeisenä tavoitteena on
Henkiset kilpailut / Cultural competitions
 Ilmoittautuminen Australasian Suomalaisten Liiton (ASL) Suomi-Päivien kilpailuihin Registration for Australasian Federation of Finnish Societies and Clubs (AFFSC) Finnish Festival competitions (Huom. tekstaa
Ilmoittautuminen Australasian Suomalaisten Liiton (ASL) Suomi-Päivien kilpailuihin Registration for Australasian Federation of Finnish Societies and Clubs (AFFSC) Finnish Festival competitions (Huom. tekstaa
Tarua vai totta: sähkön vähittäismarkkina ei toimi? 11.2.2015 Satu Viljainen Professori, sähkömarkkinat
 Tarua vai totta: sähkön vähittäismarkkina ei toimi? 11.2.2015 Satu Viljainen Professori, sähkömarkkinat Esityksen sisältö: 1. EU:n energiapolitiikka on se, joka ei toimi 2. Mihin perustuu väite, etteivät
Tarua vai totta: sähkön vähittäismarkkina ei toimi? 11.2.2015 Satu Viljainen Professori, sähkömarkkinat Esityksen sisältö: 1. EU:n energiapolitiikka on se, joka ei toimi 2. Mihin perustuu väite, etteivät
VAASAN YLIOPISTO Humanististen tieteiden kandidaatin tutkinto / Filosofian maisterin tutkinto
 VAASAN YLIOPISTO Humanististen tieteiden kandidaatin tutkinto / Filosofian maisterin tutkinto Tämän viestinnän, nykysuomen ja englannin kandidaattiohjelman valintakokeen avulla Arvioidaan viestintävalmiuksia,
VAASAN YLIOPISTO Humanististen tieteiden kandidaatin tutkinto / Filosofian maisterin tutkinto Tämän viestinnän, nykysuomen ja englannin kandidaattiohjelman valintakokeen avulla Arvioidaan viestintävalmiuksia,
Asiakaspalautteen merkitys laboratoriovirheiden paljastamisessa. Taustaa
 Asiakaspalautteen merkitys laboratoriovirheiden paljastamisessa Paula Oja, TtT Laboratorio, Oulun yliopistollinen sairaala Potilasturvallisuustutkimuksen päivät 26. 27.1.2011 1 Taustaa Laboratorion tulee
Asiakaspalautteen merkitys laboratoriovirheiden paljastamisessa Paula Oja, TtT Laboratorio, Oulun yliopistollinen sairaala Potilasturvallisuustutkimuksen päivät 26. 27.1.2011 1 Taustaa Laboratorion tulee
Vakiomallisten kuumakanavajärjestelmien kytkeminen Wiring of standard hotrunner systems
 Teknillinen dokumentaatio Technical documentation E. Vakiomallisten kuumakanavajärjestelmien kytkeminen Wiring of standard hotrunner systems Version.0 ! Tärkeä turvaohje / Important safety instruction
Teknillinen dokumentaatio Technical documentation E. Vakiomallisten kuumakanavajärjestelmien kytkeminen Wiring of standard hotrunner systems Version.0 ! Tärkeä turvaohje / Important safety instruction
