Title: Enhancement of protein production via the strong DIT1 terminator and two RNA-binding proteins in Saccharomyces cerevisiae
|
|
|
- Aleksi Salo
- 6 vuotta sitten
- Katselukertoja:
Transkriptio
1 Title: Enhancement of protein production via the strong DIT1 terminator and two RNA-binding proteins in Saccharomyces cerevisiae Authors: Yoichiro Ito, Takao Kitagawa, Mamoru Yamanishi, Satoshi Katahira, Shingo Izawa, Kenji Irie, Makoto Furutani-Seiki and Takashi Matsuyama Supplementary Figures S1-S8 and Tables S1-S5.
2 Yeast genomic tiling collection pool X DIT1t strain TDH3pro GFP DIT1t X Y Z gene X gene Y gene Z Supplymentary Figure S1. Scheme of screening for DIT1t-activating factors by using the Yeast Genomic Tiling Collection. The DIT1t strain, which expresses GFP under the control of the TDH3 promoter (TDH3pro) and the DIT1 terminator (DIT1t), was transformed with plasmid DNAs isolated from the yeast genomic tiling collection. The GFP fluorescence intensity of the transformants was then analyzed by flow cytometry and transformants with the highest GFP fluorescence intensity (marked with grey shading) were selected for further study. Plasmid DNAs containing multiple genes (marked as genes X, Y, Z) were then isolated from these transformants, and individual candidate genes (e.g., gene X) were cloned by gap-repair complementation, resulting in identification of the enhancing genes for the DIT1 terminator.
3 GFP/SS Log phase * * * * A43410LogA A43410LogA A43410LogA A43410LogA A43410LogA A43410LogA A43410LogB A43410LogB A43410LogB A43410LogB A43410LogB A43410LogB A43410LogC A43410LogC A43410LogC A43410LogC A43410LogC A43410LogC A43410LogD A43410LogD A43410LogD A43410LogD A43410LogD A43410LogD A43410LogE A43410LogE A43410LogE A43410LogE A43410LogE A43410LogE A43410LogF A43410LogF A43410LogF A43410LogF A43410LogF A43410LogF A43410LogG A43410LogG A43410LogG A43410LogG A43410LogG A43410LogG A43410LogH A43410LogH A43410LogH A43410LogH A43410LogH A43410LogH GFP/SS Log phase A43416LogA A43416LogA A43416LogA A43416LogA A43416LogA A43416LogA A43416LogB A43416LogB A43416LogB A43416LogB A43416LogB A43416LogB A43416LogC A43416LogC A43416LogC A43416LogC A43416LogC A43416LogC A43416LogD A43416LogD A43416LogD A43416LogD A43416LogD A43416LogD A43416LogE A43416LogE A43416LogE A43416LogE A43416LogE A43416LogE A43416LogF A43416LogF A43416LogF A43416LogF A43416LogF A43416LogF A43416LogG A43416LogG A43416LogG A43416LogG A43416LogG A43416LogG A43416LogH A43416LogH A43416LogH A43416LogH A43416LogH A43416LogH Supplementary Figure S2. Raw data used in the first screening. Candidate clones were located in the upper-row data set. No candidate clone was detected in the lower-row data set..
4 12 * * Flruorescence Intensity (a.u.) 6 * * 0 pgp564 7F9 10A10 11E10 12B3 Supplementary Figure S3. Effects of plasmids on the DIT1t strain. DNA of each plasmid vector pgp564 (Control), 7F9, 10A10, 11E10, or 12B3 (Extended Data Table 1) was transformed into the DIT1t strain. GFP fluorescence intensities of transformants were measured by flow cytometry. Three or four independent experiments were performed. Error bars are standard deviations. *, P < 0.01 by Student s t-test compared with Control.
5 A 15 B 15 Flruorescence Intensity (a.u.) 10 5 * control NAB6 overexpression Flruorescence Intensity (a.u.) 10 5 * control PAP1 overexpression 0 DIT1t RPL41Bt RPL15At RPL3t IDP1t PGK1t 0 DIT1t RPL41Bt RPL15At RPL3t IDP1t PGK1t C 15 D 15 control control Flruorescence Intensity (a.u.) 10 5 * MOS2 overexpression Flruorescence Intensity (a.u.) 10 5 * * CCC1 overexpression * * * * 0 DIT1t RPL41Bt RPL15At RPL3t IDP1t PGK1t 0 DIT1t RPL41Bt RPL15At RPL3t IDP1t PGK1t Supplementary Figure S4. Effects of enhancement of the top-five-ranked terminators. (A) Strains DIT1t, RPL41Bt, RPL15At, RPL3t, IDP1t, and PGK1t harboring GFP under the control of the TDH3 promoter and each terminator were transformed with (A) pgp564 or pgp564-nab6 vector; (B) pgp564 or pgp564-pap1 vector; (C) pgp564 or pgp564-mos2 vector; or (D) pgp564 or pgp564-ccc1 vector. The resulting GFP fluorescence intensities were measured by flow cytometry. Three or four independent experiments were performed. Error bars are standard deviations. *, P < 0.01 by Student s t-test compared with pgp564.
6 10 NAB6 Flruorescence Intensity (a.u.) 5 * nab6 0 DIT1t RPL41Bt RPL15At RPL3t IDP1t PGK1t Supplementary Figure S5. Effects of NAB6 deletion on the activities of the top-five-ranked terminators. Strains DIT1t, RPL41Bt, RPL15At, RPL3t, IDP1t, and PGK1t harboring GFP under the control of the TDH3 promoter. The resulting GFP fluorescence intensities were measured by flow cytometry. Four independent experiments were performed. Error bars are standard deviations. *, P < 0.01 by Student s t-test compared between NAB6 and nab6 background.
7 A UAAAGUAAGAGCGCUACAUU GGUCUACCUUUUUGUUCUUU UACUUAAACAUUAGUUAGUU CGUUUUCUUUUUCUCAUUUU d1 d2 UUUAUGUUUCCCCCCCAAAG UUCUGAUUUUAUAAUAUUUU AUUUCACACAATTCCAUUUA ACAGAGGGGGAATAGAUUCU d3 d4 d5 UUAGCUUAGAAAAUUAGUGA UCAAUAUAUAUUUGCCUUUC UUUUCAUC d6 B 16 Fluorescent Intensity (a.u.) 8 Control NAB6 overexpression 0 WT d1 d2 d3 d4 d5 d6 PAP1 overexpression Supplementary Figure S6. Deletion analysis of the DIT1 terminator for identification of the Pap1p- and Nab6p-recruiting region. (A) Sequence of the DIT1 3 -UTR. Deletion sites (d1 to d6) are underlined. (B) Strains harboring the DIT1 terminator region (Control) with deletion of d1 to d6 were constructed and then transformed with pgp564 (white), pgp564-nab6 (gray), or pgp564-pap1 (blue). The GFP fluorescence intensities were measured by flow cytometry. Three independents experiments were performed. Error bars are standard deviations.
8 PGK1t DIT1t-inserted PGK1t GATCA TTTTTTTCTT GATCAAAAACATTAGTTAGTTCGTTTTTTTCTT PGK1t 17 TTGAA TCGATAGATCA 29 TTTTTTTCTT DIT1t-substituted PGK1t CATTAGTTAGTTCG Supplementary Figure S7. Terminator regions of DIT1t-inserted PGK1t (A) and DIT1t-substituted PGK1t (B). The position of insertion or substitution of the sequence of DIT1t is indicated above the sequence of PGK1t. The candidate cis sequence, AGTTCG, is indicated in red.
9 2.0 Relative FI 1.0 ASP3 PIR1 FMP 52 MFA1 SCW4 TIR2 YDR 133C RPL 26B RPL 26A HSP 150 YLR 285C SUC2 GPB1 YGP1 SUI1 YDL 159W DDR2 YPR 053C Control NAB6 overexpression PAP1 overexpression NAB6+PAP1 overexpression Supplementary Figure S8. Activation of terminators of Nab6p-binding mrna genes by overexpression of NAB6 and PAP1. Strains harboring each terminator region were constructed and then transformed with pgp564 (white), pgp564-nab6 (gray), pgp564-pap1 (blue), or pgp564-nab6-pap1 (green). GFP fluorescence intensities were then measured by flow cytometry. Three independent experiments were performed. Error bars are standard deviations.
10 Supplementary Table S1. Genes included in genomic locus Plasmid name Chomosome location Genes in the genomic locus 7F9 chr k-960k 10A10, 10B2, 10D6, 10F2 chr k-450k 11E10 chr k-583k 12B3, 12B4 chr k-38k *: Asterisk shows enhanced genes for the DIT1 terminator YHB1, MOS2 *, SPG1, YGR237C, KEL2, PEX2 AUR1, MRP17, DID4, MET14, VPS1, PAP1 *, ECM9, YKR005C MSC3, CCC1 *, RSA3, UTP13, YLR222C-A YML119W, NGL3, NAB6 *, YML116W-A
11 Supplementary Table S2. DNA Sequences of terminator regions. Name d1 d2 d3 d4 d5 d6 d7 d8 d9 DNA sequence TAAAGTAAGAGCGCTACATTTTTGTTCTTTTACTTAAACATTAGTTAGTT CGTTTTCTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAGTTCTGATTTT ATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGGAATAGATTCT TTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTCTTTTCATCTT TTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT CGTTTTCTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAGTTCTGATTTT ATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGGAATAGATTCT TTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTCTTTTCATCTT TTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT TTAGTTAGTTCGTTTTCTTTTTCTCATTTTCCCCCCAAAGTTCTGATTTT ATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGGAATAGATTCT TTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTCTTTTCATCTT TTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT TTAGTTAGTTCGTTTTCTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAG TTCTGATTTTATTTCACACAATTCCATTTAACAGAGGGGGAATAGATTCT TTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTCTTTTCATCTT TTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT TTAGTTAGTTCGTTTTCTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAG TTCTGATTTTATAATATTTTATTTCACACAATTCCATTTAAATAGATTCT TTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTCTTTTCATCTT TTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT TTAGTTAGTTCGTTTTCTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAG TTCTGATTTTATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGG AATAGATTCTTTAGCTTAGATCAATATATATTTGCCTTTCTTTTCATCTT TTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT TAAAGTAAGAGCGCTACATTGGTCTACCTTTTTCTTTTACTTAAACATTA GTTAGTTCGTTTTCTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAGTTC TGATTTTATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGGAAT AGATTCTTTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTCTTT TCATCTTTTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT TAAAGTAAGAGCGCTACATTGGTCTACCTTTTTGTTTTACTTAAACATTA GTTAGTTCGTTTTCTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAGTTC TGATTTTATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGGAAT AGATTCTTTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTCTTT TCATCTTTTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT TAAAGTAAGAGCGCTACATTGGTCTACCTTTTTGTTCTACTTAAACATTA GTTAGTTCGTTTTCTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAGTTC TGATTTTATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGGAAT AGATTCTTTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTCTTT TCATCTTTTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT
12 d10 TAAAGTAAGAGCGCTACATTGGTCTACCTTTTTGTTCTTTTTAAACATTA GTTAGTTCGTTTTCTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAGTTC TGATTTTATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGGAAT AGATTCTTTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTCTTT TCATCTTTTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT d11 TAAAGTAAGAGCGCTACATTGGTCTACCTTTTTGTTCTTTTACTACATTA GTTAGTTCGTTTTCTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAGTTC TGATTTTATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGGAAT AGATTCTTTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTCTTT TCATCTTTTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT d12 TAAAGTAAGAGCGCTACATTGGTCTACCTTTTTGTTCTTTTACTTAATTA GTTAGTTCGTTTTCTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAGTTC TGATTTTATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGGAAT AGATTCTTTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTCTTT TCATCTTTTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT d13 GTTAGTTCGTTTTCTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAGTTC TGATTTTATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGGAAT AGATTCTTTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTCTTT TCATCTTTTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT d14 TTAAGTTCGTTTTCTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAGTTC TGATTTTATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGGAAT AGATTCTTTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTCTTT TCATCTTTTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT d15 TTAGTTTCGTTTTCTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAGTTC TGATTTTATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGGAAT AGATTCTTTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTCTTT TCATCTTTTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT d16 TTAGTTAGTTTTTCTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAGTTC TGATTTTATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGGAAT AGATTCTTTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTCTTT TCATCTTTTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT d17 TTAGTTAGTTCGTCTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAGTTC TGATTTTATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGGAAT AGATTCTTTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTCTTT TCATCTTTTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT d18 TTAGTTAGTTCGTTTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAGTTC TGATTTTATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGGAAT AGATTCTTTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTCTTT TCATCTTTTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT d19 TTAGTTAGTTCGTTTTCTTCTCATTTTTTTATGTTTCCCCCCCAAAGTTC TGATTTTATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGGAAT AGATTCTTTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTCTTT TCATCTTTTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT
13 d20 d21 TTAGTTAGTTCGTTTTCTTTTCATTTTTTTATGTTTCCCCCCCAAAGTTC TGATTTTATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGGAAT AGATTCTTTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTCTTT TCATCTTTTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT TTAGTTAGTTCGTTTTCTTTTTCTTTTTTTATGTTTCCCCCCCAAAGTTC TGATTTTATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGGAAT AGATTCTTTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTCTTT TCATCTTTTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT
14 Name m1 m2 m3 m4 m5 m6 m7 m8 m9 m10 m11 DNA sequence TTAGTTgGTTCGTTTTCTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAG TTCTGATTTTATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGG AATAGATTCTTTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTC TTTTCATCTTTTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT TTAGTTcGTTCGTTTTCTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAG TTCTGATTTTATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGG AATAGATTCTTTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTC TTTTCATCTTTTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT TTAGTTtGTTCGTTTTCTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAG TTCTGATTTTATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGG AATAGATTCTTTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTC TTTTCATCTTTTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT TTAGTTAaTTCGTTTTCTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAG TTCTGATTTTATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGG AATAGATTCTTTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTC TTTTCATCTTTTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT TTAGTTAcTTCGTTTTCTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAG TTCTGATTTTATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGG AATAGATTCTTTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTC TTTTCATCTTTTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT TTAGTTAtTTCGTTTTCTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAG TTCTGATTTTATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGG AATAGATTCTTTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTC TTTTCATCTTTTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT TTAGTTAGaTCGTTTTCTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAG TTCTGATTTTATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGG AATAGATTCTTTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTC TTTTCATCTTTTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT TTAGTTAGgTCGTTTTCTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAG TTCTGATTTTATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGG AATAGATTCTTTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTC TTTTCATCTTTTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT TTAGTTAGcTCGTTTTCTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAG TTCTGATTTTATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGG AATAGATTCTTTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTC TTTTCATCTTTTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT TTAGTTAGTaCGTTTTCTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAG TTCTGATTTTATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGG AATAGATTCTTTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTC TTTTCATCTTTTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT TTAGTTAGTgCGTTTTCTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAG TTCTGATTTTATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGG AATAGATTCTTTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTC TTTTCATCTTTTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT Position of the mutated nucleotide The 5th A to G in "AGTTAGTTCG" The 5th A to C in "AGTTAGTTCG" The 5th A to T in "AGTTAGTTCG" The 6th G to A in "AGTTAGTTCG" The 6th G to C in "AGTTAGTTCG" The 6th G to T in "AGTTAGTTAG" The 7th T to A in "AGTTAGTTAG" The 7th T to G in "AGTTAGTTAG" The 7th T to C in "AGTTAGTTCG" The 8th T to A in "AGTTAGTTAG" The 8th T to G in "AGTTAGTTAG"
15 m12 m13 m14 m15 m16 m17 m18 m19 m20 m21 m22 TTAGTTAGTcCGTTTTCTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAG TTCTGATTTTATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGG AATAGATTCTTTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTC TTTTCATCTTTTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT TTAGTTAGTTaGTTTTCTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAG TTCTGATTTTATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGG AATAGATTCTTTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTC TTTTCATCTTTTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT TTAGTTAGTTgGTTTTCTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAG TTCTGATTTTATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGG AATAGATTCTTTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTC TTTTCATCTTTTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT TTAGTTAGTTtGTTTTCTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAG TTCTGATTTTATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGG AATAGATTCTTTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTC TTTTCATCTTTTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT TTAGTTAGTTCaTTTTCTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAG TTCTGATTTTATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGG AATAGATTCTTTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTC TTTTCATCTTTTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT TTAGTTAGTTCcTTTTCTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAG TTCTGATTTTATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGG AATAGATTCTTTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTC TTTTCATCTTTTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT TTAGTTAGTTCtTTTTCTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAG TTCTGATTTTATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGG AATAGATTCTTTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTC TTTTCATCTTTTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT TTgGTTAGTTCGTTTTCTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAG TTCTGATTTTATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGG AATAGATTCTTTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTC TTTTCATCTTTTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT TTcGTTAGTTCGTTTTCTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAG TTCTGATTTTATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGG AATAGATTCTTTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTC TTTTCATCTTTTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT TTtGTTAGTTCGTTTTCTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAG TTCTGATTTTATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGG AATAGATTCTTTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTC TTTTCATCTTTTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT TTAaTTAGTTCGTTTTCTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAG TTCTGATTTTATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGG AATAGATTCTTTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTC TTTTCATCTTTTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT The 8th T to C in "AGTTAGTTAG" The 9th C to A in "AGTTAGTTCG" The 9th C to G in "AGTTAGTTCG" The 9th C to T in "AGTTAGTTCG" The 10th G to A in "AGTTAGTTCG" The 10th G to C in "AGTTAGTTCG" The 10th G to T in "AGTTAGTTCG" The 1st A to G in "AGTTAGTTCG" The 1st A to C in "AGTTAGTTCG" The 1st A to T in "AGTTAGTTCG" The 2nd G to A in "AGTTAGTTCG"
16 m23 m24 m25 m26 m27 m28 m29 m30 TTAcTTAGTTCGTTTTCTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAG TTCTGATTTTATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGG AATAGATTCTTTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTC TTTTCATCTTTTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT TTAtTTAGTTCGTTTTCTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAG TTCTGATTTTATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGG AATAGATTCTTTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTC TTTTCATCTTTTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT TTAGaTAGTTCGTTTTCTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAG TTCTGATTTTATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGG AATAGATTCTTTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTC TTTTCATCTTTTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT TTAGgTAGTTCGTTTTCTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAG TTCTGATTTTATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGG AATAGATTCTTTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTC TTTTCATCTTTTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT TTAGcTAGTTCGTTTTCTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAG TTCTGATTTTATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGG AATAGATTCTTTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTC TTTTCATCTTTTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT TTAGTaAGTTCGTTTTCTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAG TTCTGATTTTATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGG AATAGATTCTTTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTC TTTTCATCTTTTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT TTAGTgAGTTCGTTTTCTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAG TTCTGATTTTATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGG AATAGATTCTTTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTC TTTTCATCTTTTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT TTAGTcAGTTCGTTTTCTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAG TTCTGATTTTATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGG AATAGATTCTTTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTC TTTTCATCTTTTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT The 2nd G to C in "AGTTAGTTCG" The 2nd G to T in "AGTTAGTTCG" The 3rd T to A in "AGTTAGTTCG" The 3rd T to G in "AGTTAGTTCG" The 3rd T to C in "AGTTAGTTCG" The 4th T to A in "AGTTAGTTCG" The 4th T to G in "AGTTAGTTCG" The 4th T to C in "AGTTAGTTCG"
17 Name DNA sequence length DIT1t TTAGTTAGTTCGTTTTCTTTTTCTCATTTTTTTATGTTTCCCCCCCAAAG TTCTGATTTTATAATATTTTATTTCACACAATTCCATTTAACAGAGGGGG AATAGATTCTTTAGCTTAGAAAATTAGTGATCAATATATATTTGCCTTTC TTTTCATCTTTTCAGTGATATTAATGGTTTCGAGACACTGCAATGGCCCT 250 bp PGK1t DIT1t-inserted PGK1t DIT1tsubstituted PGK1t ATTGAATTGAATTGAAATCGATAGATCAATTTTTTTCTTTTCTCTTTCCC CATCCTTTACGCTAAAATAATAGTTTATTTTATTTTTTGAATATTTTTTA TTTATATACGTATATATAGACTATTATTTATCTTTTAATGATTATTAAGA TTTTTATTAAAAAAAAATTCGCTCCTCTTTTAATGCCTTTATGCAGTTTT TTTTTCCCATTCGATATTTCTATGTTCGGGTTCAGCGTATTTTAAGT ATTGAATTGAATTGAAATCGATAGATCAAAAACATTAGTTAGTTCGTTTT TTTCTTTTCTCTTTCCCCATCCTTTACGCTAAAATAATAGTTTATTTTAT TTTTTGAATATTTTTTATTTATATACGTATATATAGACTATTATTTATCT TTTAATGATTATTAAGATTTTTATTAAAAAAAAATTCGCTCCTCTTTTAA TGCCTTTATGCAGTTTTTTTTTCCCATTCGATATTTCTATGTTCGGGTTC AGCGTATTTTAAGT ATTGAATTGAATTGAAACATTAGTTAGTTCGTTTTTTTCTTTTCTCTTTC CCCATCCTTTACGCTAAAATAATAGTTTATTTTATTTTTTGAATATTTTT TATTTATATACGTATATATAGACTATTATTTATCTTTTAATGATTATTAA GATTTTTATTAAAAAAAAATTCGCTCCTCTTTTAATGCCTTTATGCAGTT TTTTTTTCCCATTCGATATTTCTATGTTCGGGTTCAGCGTATTTTAAGT 247 bp 264 bp 249 bp
18 Supplementary Table S3. List of genes, pimers and amplified terminator regions of NAB6 -binding mrna genes. Systematic Name Gene Name 5'-primer 3'-primer Sequence of amplified region 1 YLR155C ASP3-1 AGCCTGATCAGTGACTTTTCATC GCGGCCATATCTACCAGAAA AGCCTGATCAGTGACTTTTCATCACACTGTGTTTTTATATAGTTCTTAGTAGTAAATATACTCTATAATTTCTACATCCTTTTC ATGATGTGCCAGTTAAGCTATTTTTAGCAAACAAAAAGATGAATTTGAGTATTGAGCACATCATTCACTGTTCAGCTGATACAA AACATATGATTTCTAATATTCACATAGGAAAAATCGCTCCAAACATCGAAGTATCATCGCTACTTTGGTCTTTTATCTGCTTCT CTTCTCTTCGCTCTTTTCACTGCTCGTGCCGCTGCAACGCTCCAACTCAGTTCTATCTCAGCTCCTTCCCCTTATCCCTTGAGC AGTGTCAAGGCGAGGCTGCGTGATGTATTCGTGCGCGAAAAAAAAAACAACTGCAGCACCTGAGTTTCGCGTATGGTCACCCAC TACACTACTCGGTCAGGCTCTTACCAGCTTAACTACAGTTGATCGGACGGGAAACGGTGCTTTCTGGTAGATATGGCCGC 2 YKL164C PIR1 TCATAAGTTTACCTCTCATAGTCGCA GGCTCCACGGTACACAAAGT TCATAAGTTTACCTCTCATAGTCGCATGAAAACATAAAGTCATCGTAGCATATTCTGTACATTAGTCATTTCTGTCTCCAAAGG GCATAATTTGGGTCGGTTCATTTTTTTCTTCTTAATTCGTCTCTTCATTTCATAAAGCTTTCTTTTGAAACCTATGAAATGAGC TTTACAGTTCTTCACTCTTTTTTTTAAAGTTTGTTTATTTTTTTATACTATATACATTATTATTTCTAAAAAATCATTATAAAT TACGTTAAAAGCGCACCTTATACTTTGATAGAGTTTTCCCAGAAAAAAAACCAACGAATTTGATAGTACCAAGACTTCACGGGA TTGAAATCGGATGCAACTTCAACTATTCCTGGGACAGAATATTTCTCAGACATTACTTTGTGTACCGTGGAGCC 3 YER004W FMP52 ATCAACCAACGGCTTCTTCT TGTGAGTCCTGGTTTGTTGC ATCAACCAACGGCTTCTTCTCGCAGCTACAGTTGTATTTTACTGTGACGTACTTTTGCGCGCGTACGGGTTCAATGAATTACGG ATTTTCAACAACTTCACACAATCATAGCTTTTTTTACTTTTTCGCCTCATTATTTTCTTTAAAGAAACGATGATAAACAGAGAT TTATCGTCATGTTATTTCGTTCGTATATATCGTATTTGCGTTTTAGCTGTATAAAGTAAAATACTATATCTAACGTGGCTTTTT TTATTTCCCCGTCTGCCCTCTTATGCTCATAGAAAACACTAATGATCGGTTTGGTATCGTCATAGATGCAGGGTCTTCGGGTTC GAGAATCCATGTGTTTAAGTGGCAGGACACGGAATCGCTTCTTCATGCAACAAACCAGGACTCACA 4 YDR461W MFA1 GTTATTGCTTAGTTTCTGCGTACA TAGATCCGCCCTCTTACGAA 5 YGR279C SCW4 GTCTACTCAATTGTAAAAAAAAACAAA CCAAGAGGATCGAGAAACCA GTTATTGCTTAGTTTCTGCGTACAAAAACGTTGTTCTCCCTCCTTTATCTTCCTTTTCCGCTACACCAATATATCATGTTTGTT CGTAATATTTCTTTTTAGACCTAATAATAAATATCCTAAGTAACTATATTATATAAAATATTTTGATACCCTGTACCTGCTTTT GTTATCGTTGTACATCCATGCACACGCTCATATATATATATATATATATATATATGTATATGTACATATACCGCTTACCACCTA CCGTGAAGTATATGTAAGGGTCTTCGCACCCGGATATCCCTTGTGGGATCTTGGATGCGGATGGTGAGTGTAAACAGGCCTCAT AAAGCTGTCTCTGTGTGCGGAAGATGTCGTTTCATCGGCTTCGACCTCCTCCTTATCTTGCTTCATTGTTTCTTCGTAAGAGGG CGGATCTA GTCTACTCAATTGTAAAAAAAAACAAAAAAAATCTTGATTTAAAAAACGATACCCAATATTTTTAACTTTAAATTAAGGCTTTG ATAATCTTCTTTTTCTCTGTTATATCAAAGGACTGCTTTCTGGTTTGCCTTCGCTAATCTTTTTTTTTTTTTTTCATTTTCCTA TTTTAGTTCGTTTATGATCTCATTTGTATTTCTGTATATTTGTCTTAATACGAAATACGAGTTTTTATTTTATATTCTACCTTT CTTTTTTTTTTGGCTAAAAGCGTGTACGGTTCTATGTAAATATGTACGATTACATGTGTAATGTTTGTTTGGCCGGGCTCTTCA TCTATGCCGTTGCCTTCTTGCAGGCGGAGTCCTGGATCTGTTACGATGGTTTCTCGATCCTCTTGG 6 YOR010C TIR2 TTTTAAATGGAATTTTTAATGTTCG GCCGCCATGCTACTATGATT TTTTAAATGGAATTTTTAATGTTCGTATATACCAATTTATCTATTGAATAGTTTTTTTTAAATTTTACTGTTTCTTTTAAACCT CGAATTATTGTGAGAACTGATTTCATCAACTGGAGGCTGTAGTGTACGTTTTGAAAACGGAAATGATTTACCGTTTTCTTTTAT GATGATGCTAGGATAGGCACCTTCAAACATGCATGTCTTCCTGCCCACATTCTGAAAATATTTAACGATGGATGAAAGAAATCA ACAATAGAAAAGGCCAGTTGTCAGTTTTTATATAATACAGAGAAGTACATATGACAGTTTAAAAGTTAAGCATTAACTGGTAAG TTGAAATAATAATATAATCATAAGCGGAACGAACATTTTCGACACGTACTAAAATCATAGTAGCATGGCGGC 7 YDR133C YDR133C TTTCTTATTCTAATTCTAGG CGAGAGGAGAATCCATTAG 8 YGR034W RPL26B TTGGAATAAACTAGCTCGACTTTAAC TGTGGCCAATACTGCTTCAG 9 YLR344W RPL26A ATTATGTAATTCCCTGAAAACTTCC TTTACCCACCTTTTGACGGT 10 YJL159W HSP150 TAGTTCTTTTATCCTGATGACTTTTTC AATAGACGTAGCCACATGCAG TTTCTTATTCTAATTCTAGGAGGCAACACTCCATAACTAAATGTTTATTATAATAATATTAATATAAAGTTAATATCTCTTAGG TTACACGCAACTAAACTTTTTTTATATATACTGGTTGCATTGTATTTTTATCATATATCTTAATATAGGTTGTTTAAGGTTCGT TAATTCAAAAATAATGTTTTTTTGTTTGCCCAGTCGGTTTACGCTAATCAAGAACATGGATTCAATATCTTACATAATAAGCGG ACGATGAGTCTATCTCGGTTTCCTCTTTTCTGAAATATAATAGTTTTATATTAGTGATTCATTATTACGATCTGGCCGGCTTAT CATTGCTTCTTTTACAAACGTATCCTTTTAAGATTTTATCCGGACCACTGAGCTGGCATTTTTCTTAGCTTGCTTTTGTCTTTG TTGCACGGGCGACTGAACTTCTTGGAAAATCATGTAAGCCGCAATTGCTTACTAAGTCAAAATTCTAATGGATTCTCCTCTCG TTGGAATAAACTAGCTCGACTTTAACATAGATCATACAAAAAATACAATAAATAAATCCTGTGTTTCTTTTATGTATATTTTTT ATCAATTTCAATTTAGTTTAGTTTAGTTTCTTTAAAAATAAAATTGTAATTTCTCACATTCTGCCTTCGGGTCTCAAGGGCTGG TTGGTAGACGAATGCGCTTGTTTCTAGCTGAGCGGCAGATGCATGAATTCAATGCAAGAAAATGAGAAAACTTCCAATATTAAA ATGTGATTGTTAATTATTAGTATATTTATGACAATATATATATGTACATAGGGTGATTATGAAGATGAGTAAGATGCGAATGGA TATGAGGATATGAATCTTTTTAGAGACAATGAAGATGCAAAGAAGTAGTTAAGCACATTGACCTCAGGGTTGCTATATGCTACT TTTAGTCCGTTGGATTTCTGAAGCAGTATTGGCCACA ATTATGTAATTCCCTGAAAACTTCCTCGAAATTTATTGCACTTTTTTTACTCAAACCATTTAATAATCATATGTAAAACGTATA AAATAGTATCCATTCCACTGTACTCAGTGTCTTACGTAAAGGGCTGTATACTGGCACTACCTTTTACTATGGATATGTGCTAGC TTTTTCGGAGAAGAAATGTTCGCCTTACGCTAGAAACCGGGGTTAAGGGGGCGTGAGAATGAATATTGGCAGACAAGAACCGAG ATAGAATCTTAACACAAAAACAGTTCATCCTACTTCTACGCCTTTATACAGTTATACTGAAAGATCTTTGCATAGGGGCATAGT TTAAAATAAAATATAGGGTTGTCATAGCAAACATGCCAAATGTGCTTTCTGATGATGAAGAGCTCCTTAATGGACTAGGAAGTG AGATTATGAAACCCTCTAGACAAGGTAACCATATGGCAAGGACCGTCAAAAGGTGGGTAAA TAGTTCTTTTATCCTGATGACTTTTTCTCATTTGCATTGATTAGAAAGGAAAAAAAGAAGTGTCCTTTTCTACTACTACTCTAG TCGCATCCATTCCTTTGCATTTATCTTTTCTGCGGTTGGCCAATCCATTCTTCCGAGAATTTGGCTAGCCATACTTGATGTTTT CCCATTATTGGTTCGTTTGGCAATGCTAATTTTCTTAATTGCCCCTTATATACTCTTCCATAAAATGTTTTTTTTATAACTAAT TTTCTGTATATCATTATCTAATAATCTTATAAAATGTTAAAAAGACTTGGAAAGCAACGAGTGATCGTGACCACATAATTGCCT CGCTACACGGCAAAAATAAGCCAGTCCTAATGTGTATATTAAAGGCTGCATGTGGCTACGTCTATT 11 YLR285C-A YLR285C-A TGAGAATGCCAATTCCGC TGGTTGTTTCAACTGATTATCCA 12 YIL162W SUC2 AGGTTATAAAACTTATTGTCTTTTTT GCTTGGTATCCATTTCCCTC 13 YOR371C GPB1 TAATTAATTCCGCGGGGC TGAAATGTTGCTGTTAAACCCTT 14 YNL160W YGP1 AGATATTAAATCTAGCGAAGCATAGAG GACACAAAAGAAGGGGGTGA 15 YNL244C SUI1 GTTCAAGGCTTACGCCGAGT CATTCTTGAGAGTCTCCGGC 16 YDL159W-A YDL159W-A TCACTGTCCTCGTTATAAGTGATTC AAAACAGGTTCATTTTGCGG 17 YOL052C-A DDR2 GAAAAACGCCGCTTACTGC CTTTGGTTTCATGGCGAAAT TGAGAATGCCAATTCCGCCCACTTACTACTTTTCATGTGTTAAATAGTACGCATAAACACGAGATCTGACTTCGATTGTTTCCT CGTCTTCGTCAAACATCGGTTTCCAATTAACCATTTCAATTAGCTGAGGAACCAAATGGAACTCGTTCTTAGCCAATTCGAAGA ATTCTAAATCCTTCTCCAACAATTTTGGTCTATGCGGTGAAAATACTACCAACGCCTGACCTTTCTCAGCTAATAAATCCTTTG TTGTTTGAAGTAATTTGTGATGTTCCGTATGATTGAAGACTAAATCACTTAAAATGATTAAGTCAAACTTTCCATTATTGTTAC CTATTTTTTCGATATGTGCCAACAATGGAGAATAATCGTTTCCCCAAATATAACCCTCCGTACTGACATTATTAAAATCTTCAG GAACGTTAGACTTTATATTATAATCGATGTTCTGCATTAAATCAGGATCTGGATAATCAGTTGAAACAACCA AGGTTATAAAACTTATTGTCTTTTTTATTTTTTTCAAAAGCCATTCTAAAGGGCTTTAGCTAACGAGTGACGAATGTAAAACTT TATGATTTCAAAGAATACCTCCAAACCATTGAAAATGTATTTTTATTTTTATTTTCTCCCGACCCCAGTTACCTGGAATTTGTT CTTTATGTACTTTATATAAGTATAATTCTCTTAAAAATTTTTACTACTTTGCAATAGACATCATTTTTTCACGTAATAAACCCA CAATCGTAATGTAGTTGCCTTACACTACTAGGATGGACCTTTTTGCCTTTATCTGTTTTGTTACTGACACAATGAAACCGGGTA AAGTATTAGTTATGTGAAAATTTAAAAGCATTAAGTAGAAGTATACCATATTGTAAAAAAAAAAAGCGTTGTCTTCTACGTAAA AGTGTTCTCAAAAAGAAGTAGTGAGGGAAATGGATACCAAGC TAATTAATTCCGCGGGGCGGGGTTCGGATAATCCTTATGTGACAAGAGTGACTAAAGGAAAACGAGAAAATTTTTTCGTGGAAC TTGAAAAAAAAAAAAAAAAAAAAAAAGAAAACAGAACAAGTGAAGAAGATAAAAAAAGGTTCTATATCGCACTATACAACAATA GTAGCGTTTCCCGTGTTCTTTTCCCGTCATTAAATATATTCAAAAGTTGTTGCACATTTGATTTTACAGAAGTAATATACACTG ATTGTGCGTACGGTTAATTCGCAACTAAATTCAATCAGTACAAGAAAAATTTTTATTCCGTTCCATCATTATTGAAAGATAAAC TATCACATTGCGATAATAAACTAAAAGACACTAGGATACTGCCTTCGGATATAAACATTTAAGTTTAATTTATTACTGTTTTTT TAAGGGTTTAACAGCAACATTTCA AGATATTAAATCTAGCGAAGCATAGAGATTCTTTTTTTACCAGTCATCATAACCATTATTTTACAAAATTTCCACGCAACAGCT TTTATTTTCGTTCGTAATAGGAACCATACTTCCCATTAAAGCAGTATGATTGTTTTATACACCTTTTTATTGTTATTTAAGACA TACCCCAAATAATTAATTAATTAATTGATGAAAGAATCATATAAATTCTTGGCCATACATATTTTTTTATTATCTTTTGCTACA TTGACCATAGGTGAATATCTTCCCGGGGTGACACTCCACTGAAGTGGGAAAAAAAGAAAAGTTTTAAAACATTGCTCACCTTAA GTCTCGAAGGTCTAGCATTCACTACCTGTAAGTGTCAAGACCCCACATGTATACATGGATACAATAAAGAAACAATGTCCGTAT AATTTGTATACATTTTACTACAGATAAGGGATCTCACCCCCTTCTTTTGTGTC GTTCAAGGCTTACGCCGAGTCGCTTACTCGCTGCTCGCTTTCTTTCTTAGTGTTGTTATCCTATATAAAACGTAACTGTTTAGT TATTTAAGTCCAAGTGCATATTTACACTATAATATATAATATAACATCTCATCTTTTTTTATTTCTTTTTCTTCTTCTCCCGCC GCGGGCTTACATACAGAGATATAAGAACAAGTGGTACGTAGCATGCAGCTTCTCAAAAGAGATGAGCGTTCAGTTTTGAGTACA GGGAAGAAAGGCCAAATCGAGAGTACATTATCATGGGGTCGGGCGATTTGAATCTGTTGAAATCATGGAATCCCAAGCTTATGA AAAATAGAAAAAAAGTATGGGAGACCGAACAGGATCTAATTACCGAACAGCAGAAGCTTAACACGCGATTGAAAGAAATTGAAA AAGAACGAGAACTAAATGAACTGCTGAACGAATCGAGCAAGGATAAGCCGGAGACTCTCAAGAATG TCACTGTCCTCGTTATAAGTGATTCGTGGGGTAACGGCGATATCATACTATTATTGGCCCATCTGATGGATTAGAAGACCTACA GGTTAATGTGAAGCGACACTAATGATAAGAAAACTGCATCGGATCATATCTGTTTTTGCAGCGTGGTATATTGGTTGTTGGTCA TGTTTCAACGAAAGACTTTACAGAGAAGGAACTTGAAAGGGCTCAATCTTAACCTGCACCCAGATGTGGGCAATAATGGCCAAT TGCAGGAAAAGACAGAGACTCACCAGGGACAATCTCGAATAGAAGGCCACGTGATGTCTAACATTAATGCAATACAGAATAATA GCAACCTGTTTTTGCGAAGAGGCATAAAAAAAAAACTGACGTTGGATGCGTTTGGTGATGACCAAGCTATATCGAAACCAAACA CTGTGGTAATACAGCAACCGCAAAATGAACCTGTTTT GAAAAACGCCGCTTACTGCCACGATGATGATACCCTATTGACGTTTCTGAAATGTATAATTTCTTTTCTCATCTTCCCCTTTGA TATTCCATCTATAGGCCCCAGAGTAGTAAATTTGGTGCTTTAATTTTTTTTCTCTTCTTCCAGTTCGTCTCTATTTTTTCATTC CATTATATTTATTTATCAGTTTTACTTTCTCAAATATTCTTATATAACACTATTTCATCTACGTAACCGAAAATAAGTAGTTTA ATAACGATAATAATCATAATTGTGATAAAAATATTTACATTTTTCTTACTTGTCGCCCGCTTCGTTTTTGTATAGAACGTACTG TATTAGTTGTCCGTTCTTTTCACGTCTGAAGAGGTCGATTTCTTGCAATTTAACATCAACGTGGTATTTTCCATTATATACAAT CAAAGAATCATCGTCCAGTTCGATTTCGCCATGAAACCAAAG
19 18 YPR053C YPR053C CGCCAGAGGAAAAGCAGCC AACATTGCGATGAGATCATG CGCCAGAGGAAAAGCAGCCTTACGAGGCCAAGGCCCAGGCCGATAAGAAGAGATATGAATCCGAAAAGGAGTTATATAACGCCA CTTTGGCTTAGGGATACTTCCTCCCCGCGTTTTTGTCTGTGTGATTTAATTTCTTTTCCTCTTGTTCTTTTTGAAACTTGCTTT ATTATCTCTCTCCTTTCTATTCCTCTTTTCTCTACTGCCCTTTTTCTATATCTATACATATGATTAAATAAATAAATAAAAAAA TTGTATTATTAATATATATTTTATCATCATCGAGTGTTTTTCCTTTTATGTGGCCAGCCGGCCATGCATTAGGAACGTAGACCG AGTGCAGCTCTTTTCAGTCAACGGCAATATCAGTTTAAAGGCGTCCCCTTCATTCAAGTAGTAGCGGAACATCCGTTTCATTCT GATGAAACCCATTCCTTCATATAAGTTTAGAGCCGCCGAATTTTCCACCTCTGTTTCTAACATGATCTCATCGCAATGTT
20 Supplementary Table S4. Raw data sets of Figures 1 & 2. Population Genetic Background Overexpression Average log10 value (a.u.) of fluorecent intensity (FI) from 3000 cells full width at half maximum Average value of FI Mean value of Standard Deviatoin of NAB NAB NAB NAB NAB NAB6 NAB NAB6 NAB NAB6 NAB NAB6 NAB NAB6 NAB NAB6 PAP NAB6 PAP NAB6 PAP NAB6 PAP NAB6 PAP NAB6 NAB6+PAP NAB6 NAB6+PAP NAB6 NAB6+PAP NAB6 NAB6+PAP NAB6 NAB6+PAP nab nab nab nab nab nab6 PAP nab6 PAP nab6 PAP nab6 PAP nab6 PAP t-test: Paired Two Sample for Means (p<0.01) Population 1 2 significant Mean Variance Observations 5 5 Pearson Correlation df 4 t Stat P(T<=t) one-tail E-07 t Critical one-tail P(T<=t) two-tail E-06 t Critical two-tail Population 1 3 significant Mean Variance Observations 5 5 Pearson Correlation df 4 t Stat P(T<=t) one-tail E-06 t Critical one-tail P(T<=t) two-tail E-05 t Critical two-tail
21 Population 1 4 significant Mean Variance Observations 5 5 Pearson Correlation df 4 t Stat P(T<=t) one-tail E-05 t Critical one-tail P(T<=t) two-tail t Critical two-tail Population 1 5 significant Mean Variance Observations 5 5 Pearson Correlation df 4 t Stat P(T<=t) one-tail E-07 t Critical one-tail P(T<=t) two-tail E-07 t Critical two-tail significant Mean Variance Observations 5 5 Pearson Correlation df 4 t Stat P(T<=t) one-tail E-06 t Critical one-tail P(T<=t) two-tail E-06 t Critical two-tail
22 Population Ct value of GFP primer set Ct value of TUB primer set Genetic Overexpression (Sample) (Standard) Background repetition 1 repetition 2 repetition 1 repetition 2 NAB NAB NAB NAB6 NAB NAB6 NAB NAB6 NAB NAB6 PAP NAB6 PAP NAB6 PAP NAB6 NAB6+PAP NAB6 NAB6+PAP NAB6 NAB6+PAP nab nab nab nab6 PAP nab6 PAP nab6 PAP Relative GFP mrna level to TUB1 = exp(-cttub ln2)/exp(-ctgfp ln2) Population Genetic Mean of relative SD of relative Overexpression Background mrna level mrna level 1 NAB NAB6 NAB NAB6 PAP NAB6 NAB6+PAP nab nab6 PAP t-test (P<0.01) Population 1 vs 2 significant Pooled variance Test statistic P value (two-sided) E-06 Population 1 vs 3 no difference Pooled variance Test statistic 0.9 P value (two-sided) Population 1 vs 4 significant Pooled variance Test statistic 11.7 P value (two-sided) E-07 Population 1 vs 5 no difference Pooled variance Test statistic P value (two-sided) Population 1 vs 6 no difference Pooled variance Test statistic 2.7 P value (two-sided)
23 Population Genetic Background Overexpression Average log10 value (a.u.) of fluorecent intensity (FI) from 3000 cells Average value of FI Mean value of Standard Deviatoin of Relative FI (DIT1t-- Mean value of SD of relative Mean value as a standard) relative FI FI DIT1t DIT1t DIT1t DIT1t DIT1t NAB DIT1t NAB DIT1t NAB DIT1t NAB DIT1t PAP DIT1t PAP DIT1t PAP DIT1t PAP DIT1t-d DIT1t-d DIT1t-d DIT1t-d DIT1t-d2 NAB DIT1t-d2 NAB DIT1t-d2 NAB DIT1t-d2 NAB DIT1t-d2 PAP DIT1t-d2 PAP DIT1t-d2 PAP DIT1t-d2 PAP DIT1t-d DIT1t-d DIT1t-d DIT1t-d DIT1t-d7 NAB DIT1t-d7 NAB DIT1t-d7 NAB DIT1t-d7 NAB DIT1t-d7 PAP DIT1t-d7 PAP DIT1t-d7 PAP DIT1t-d7 PAP DIT1t-d DIT1t-d DIT1t-d DIT1t-d DIT1t-d8 NAB DIT1t-d8 NAB DIT1t-d8 NAB DIT1t-d8 NAB DIT1t-d8 PAP DIT1t-d8 PAP DIT1t-d8 PAP DIT1t-d8 PAP DIT1t-d DIT1t-d DIT1t-d DIT1t-d DIT1t-d9 NAB DIT1t-d9 NAB DIT1t-d9 NAB DIT1t-d9 NAB DIT1t-d9 PAP DIT1t-d9 PAP DIT1t-d9 PAP DIT1t-d9 PAP DIT1t-d DIT1t-d DIT1t-d DIT1t-d DIT1t-d10 NAB DIT1t-d10 NAB DIT1t-d10 NAB DIT1t-d10 NAB DIT1t-d10 PAP DIT1t-d10 PAP DIT1t-d10 PAP DIT1t-d10 PAP DIT1t-d DIT1t-d DIT1t-d DIT1t-d DIT1t-d11 NAB DIT1t-d11 NAB DIT1t-d11 NAB DIT1t-d11 NAB DIT1t-d11 PAP DIT1t-d11 PAP
24 DIT1t-d11 PAP DIT1t-d11 PAP DIT1t-d DIT1t-d DIT1t-d DIT1t-d12 NAB DIT1t-d12 NAB DIT1t-d12 NAB DIT1t-d12 NAB DIT1t-d12 PAP DIT1t-d12 PAP DIT1t-d12 PAP DIT1t-d12 PAP significant Mean Variance E-05 Pearson Correlation Hypothesized Mean Differe 0 df 3 t Stat P(T<=t) one-tail E-05 t Critical one-tail P(T<=t) two-tail E-05 t Critical two-tail significant Mean Variance Pearson Correlation Hypothesized Mean Differe 0 df 3 t Stat P(T<=t) one-tail E-06 t Critical one-tail P(T<=t) two-tail E-06 t Critical two-tail significant Mean Variance Pearson Correlation Hypothesized Mean Differe 0 df 3 t Stat P(T<=t) one-tail E-05 t Critical one-tail P(T<=t) two-tail E-05 t Critical two-tail
25 Population Genetic Background Overexpression Average log10 value (a.u.) of fluorecent intensity (FI) from 3000 cells Average value of FI Mean value of Standard Deviatoin of Relative FI (DIT1t-- Mean value of SD of relative Mean value as a standard) relative FI FI DIT1t DIT1t DIT1t DIT1t DIT1t NAB DIT1t NAB DIT1t NAB DIT1t NAB DIT1t PAP DIT1t PAP DIT1t PAP DIT1t PAP DIT1t-d DIT1t-d DIT1t-d DIT1t-d DIT1t-d2 NAB DIT1t-d2 NAB DIT1t-d2 NAB DIT1t-d2 NAB DIT1t-d2 PAP DIT1t-d2 PAP DIT1t-d2 PAP DIT1t-d2 PAP DIT1t-d DIT1t-d DIT1t-d DIT1t-d DIT1t-d13 NAB DIT1t-d13 NAB DIT1t-d13 NAB DIT1t-d13 NAB DIT1t-d13 PAP DIT1t-d13 PAP DIT1t-d13 PAP DIT1t-d13 PAP DIT1t-d DIT1t-d DIT1t-d DIT1t-d DIT1t-d14 NAB DIT1t-d14 NAB DIT1t-d14 NAB DIT1t-d14 NAB DIT1t-d14 PAP DIT1t-d14 PAP DIT1t-d14 PAP DIT1t-d14 PAP DIT1t-d DIT1t-d DIT1t-d DIT1t-d DIT1t-d15 NAB DIT1t-d15 NAB DIT1t-d15 NAB DIT1t-d15 NAB DIT1t-d15 PAP DIT1t-d15 PAP DIT1t-d15 PAP DIT1t-d15 PAP DIT1t-d DIT1t-d DIT1t-d DIT1t-d DIT1t-d16 NAB DIT1t-d16 NAB DIT1t-d16 NAB DIT1t-d16 NAB DIT1t-d16 PAP DIT1t-d16 PAP DIT1t-d16 PAP DIT1t-d16 PAP DIT1t-d DIT1t-d DIT1t-d DIT1t-d DIT1t-d17 NAB DIT1t-d17 NAB DIT1t-d17 NAB DIT1t-d17 NAB DIT1t-d17 PAP DIT1t-d17 PAP
26 21 DIT1t-d17 PAP DIT1t-d17 PAP significant Mean Variance Pearson Correlation Hypothesized Mean Differe 0 df 3 t Stat P(T<=t) one-tail E-05 t Critical one-tail P(T<=t) two-tail E-05 t Critical two-tail significant Mean Variance Pearson Correlation Hypothesized Mean Differe 0 df 3 t Stat P(T<=t) one-tail E-05 t Critical one-tail P(T<=t) two-tail E-05 t Critical two-tail significant Mean Variance Pearson Correlation Hypothesized Mean Differe 0 df 3 t Stat P(T<=t) one-tail E-05 t Critical one-tail P(T<=t) two-tail E-05 t Critical two-tail significant Mean Variance Pearson Correlation Hypothesized Mean Differe 0 df 3 t Stat P(T<=t) one-tail E-05 t Critical one-tail P(T<=t) two-tail t Critical two-tail significant Mean Variance Pearson Correlation Hypothesized Mean Differe 0 df 3 t Stat P(T<=t) one-tail t Critical one-tail P(T<=t) two-tail t Critical two-tail significant Mean Variance Pearson Correlation Hypothesized Mean Differe 0 df 3 t Stat P(T<=t) one-tail E-05 t Critical one-tail P(T<=t) two-tail E-05 t Critical two-tail
27 13 16 no difference Mean Variance Pearson Correlation Hypothesized Mean Differe 0 df 3 t Stat P(T<=t) one-tail t Critical one-tail P(T<=t) two-tail t Critical two-tail no difference Mean Variance Pearson Correlation Hypothesized Mean Differe 0 df 3 t Stat P(T<=t) one-tail t Critical one-tail P(T<=t) two-tail t Critical two-tail no difference Mean Variance Pearson Correlation Hypothesized Mean Differe 0 df 3 t Stat P(T<=t) one-tail t Critical one-tail P(T<=t) two-tail t Critical two-tail
28 Population Genetic Background Overexpression Average log10 value (a.u.) of fluorecent intensity (FI) from 3000 cells Average value of FI Mean value of Standard Deviatoin of Relative FI (DIT1t-- Mean value of SD of relative Mean value as a standard) relative FI FI DIT1t DIT1t DIT1t DIT1t DIT1t-d DIT1t-d DIT1t-d DIT1t-d DIT1t-d2 NAB DIT1t-d2 NAB DIT1t-d2 NAB DIT1t-d2 NAB DIT1t-d2 PAP DIT1t-d2 PAP DIT1t-d2 PAP DIT1t-d2 PAP DIT1t-d DIT1t-d DIT1t-d DIT1t-d DIT1t-d18 NAB DIT1t-d18 NAB DIT1t-d18 NAB DIT1t-d18 NAB DIT1t-d18 PAP DIT1t-d18 PAP DIT1t-d18 PAP DIT1t-d18 PAP DIT1t-d DIT1t-d DIT1t-d DIT1t-d DIT1t-d19 NAB DIT1t-d19 NAB DIT1t-d19 NAB DIT1t-d19 NAB DIT1t-d19 PAP DIT1t-d19 PAP DIT1t-d19 PAP DIT1t-d19 PAP DIT1t-d DIT1t-d DIT1t-d DIT1t-d DIT1t-d20 NAB DIT1t-d20 NAB DIT1t-d20 NAB DIT1t-d20 NAB DIT1t-d20 PAP DIT1t-d20 PAP DIT1t-d20 PAP DIT1t-d20 PAP DIT1t-d DIT1t-d DIT1t-d DIT1t-d DIT1t-d21 NAB DIT1t-d21 NAB DIT1t-d21 NAB DIT1t-d21 NAB DIT1t-d21 PAP DIT1t-d21 PAP DIT1t-d21 PAP DIT1t-d21 PAP
MALE ADULT FIBROBLAST LINE (82-6hTERT)
 Double-stranded methylation patterns of a 104-bp L1 promoter in DNAs from male and female fibroblasts, male leukocytes and female lymphoblastoid cells using hairpin-bisulfite PCR. Fifteen L1 sequences
Double-stranded methylation patterns of a 104-bp L1 promoter in DNAs from male and female fibroblasts, male leukocytes and female lymphoblastoid cells using hairpin-bisulfite PCR. Fifteen L1 sequences
FETAL FIBROBLASTS, PASSAGE 10
 Double-stranded methylation patterns of a 104-bp L1 promoter in DNAs from fetal fibroblast passages 10, 14, 17, and 22 using barcoded hairpinbisulfite PCR. Fifteen L1 sequences were analyzed for passages
Double-stranded methylation patterns of a 104-bp L1 promoter in DNAs from fetal fibroblast passages 10, 14, 17, and 22 using barcoded hairpinbisulfite PCR. Fifteen L1 sequences were analyzed for passages
tgg agg Supplementary Figure S1.
 ttaggatattcggtgaggtgatatgtctctgtttggaaatgtctccgccattaactcaag tggaaagtgtatagtaatgaatctttcaagcacacagatcacttcaaaagactgtttcaa catcacctcaggacaaaaagatgtactctcatttggatgctgtgatgccatgggtcacag attgcaattcccaagtgcccgttcttttacaccaaaatcaaagaagaatatctccccttt
ttaggatattcggtgaggtgatatgtctctgtttggaaatgtctccgccattaactcaag tggaaagtgtatagtaatgaatctttcaagcacacagatcacttcaaaagactgtttcaa catcacctcaggacaaaaagatgtactctcatttggatgctgtgatgccatgggtcacag attgcaattcccaagtgcccgttcttttacaccaaaatcaaagaagaatatctccccttt
Methods S1. Sequences relevant to the constructed strains, Related to Figures 1-6.
 Methods S1. Sequences relevant to the constructed strains, Related to Figures 1-6. A. Promoter Sequences Gal4 binding sites are highlighted in the color referenced in Figure 1A when possible. Site 1: red,
Methods S1. Sequences relevant to the constructed strains, Related to Figures 1-6. A. Promoter Sequences Gal4 binding sites are highlighted in the color referenced in Figure 1A when possible. Site 1: red,
Experimental Identification and Computational Characterization of a Novel. Extracellular Metalloproteinase Produced by Clostridium sordellii
 Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 207 Supplementary Information Experimental Identification and Computational Characterization of
Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 207 Supplementary Information Experimental Identification and Computational Characterization of
Strain or plasmid Description a Reference or source b. 168 trpc2 Laboratory. 1A780 trpc2 sigb::spc BGSC
 TABLE S1 Bacterial strains and plasmids Strain or plasmid Description a Reference or source b B. subtilis 168 trpc2 Laboratory stock 1A780 trpc2 sigb::spc BGSC BM1010 trpc2 htpx::pgs1582; Em r BM1302 trpc2
TABLE S1 Bacterial strains and plasmids Strain or plasmid Description a Reference or source b B. subtilis 168 trpc2 Laboratory stock 1A780 trpc2 sigb::spc BGSC BM1010 trpc2 htpx::pgs1582; Em r BM1302 trpc2
Plasmid Name: pmm290. Aliases: none known. Length: bp. Constructed by: Mike Moser/Cristina Swanson. Last updated: 17 August 2009
 Plasmid Name: pmm290 Aliases: none known Length: 11707 bp Constructed by: Mike Moser/Cristina Swanson Last updated: 17 August 2009 Description and application: This is a mammalian expression vector for
Plasmid Name: pmm290 Aliases: none known Length: 11707 bp Constructed by: Mike Moser/Cristina Swanson Last updated: 17 August 2009 Description and application: This is a mammalian expression vector for
Supporting information
 Supporting information Figure S1. Carotenoid biosynthesis pathway in papaya fruit which is adopted from Blas et al. (2010) (reference 4) and Nisar et al. (2015) (reference 5). Carotenoids are synthesized
Supporting information Figure S1. Carotenoid biosynthesis pathway in papaya fruit which is adopted from Blas et al. (2010) (reference 4) and Nisar et al. (2015) (reference 5). Carotenoids are synthesized
Supporting Information for
 Supporting Information for Analysis of Sogatella furcifera proteome that interact with P10 protein of Southern rice black-streaked dwarf virus Win Than*, Faliang Qin*, Wenwen Liu, Xifeng Wang ** State
Supporting Information for Analysis of Sogatella furcifera proteome that interact with P10 protein of Southern rice black-streaked dwarf virus Win Than*, Faliang Qin*, Wenwen Liu, Xifeng Wang ** State
Supplementary information: Biocatalysis on the surface of Escherichia coli: melanin pigmentation of the cell. exterior
 Supplementary information: Biocatalysis on the surface of Escherichia coli: melanin pigmentation of the cell exterior Martin Gustavsson, David Hörnström, Susanna Lundh, Jaroslav Belotserkovsky, Gen Larsson
Supplementary information: Biocatalysis on the surface of Escherichia coli: melanin pigmentation of the cell exterior Martin Gustavsson, David Hörnström, Susanna Lundh, Jaroslav Belotserkovsky, Gen Larsson
I. Principles of Pointer Year Analysis
 I. Principles of Pointer Year Analysis Fig 1. Maximum (red) and minimum (blue) pointer years. 1 Fig 2. Principle of pointer year calculation. Fig 3. Skeleton plot graph created by Kinsys/Kigraph programme.
I. Principles of Pointer Year Analysis Fig 1. Maximum (red) and minimum (blue) pointer years. 1 Fig 2. Principle of pointer year calculation. Fig 3. Skeleton plot graph created by Kinsys/Kigraph programme.
Supplementary Table S1. Material list (a) Parameters Sal to Str
 Tooth wear as a means to quantify intra-specific variations in diet and chewing movements - Scientific Reports 2016, 6:3037 Ivan Calandra, Gaëlle Labonne, Ellen Schulz-Kornas, Thomas M. Kaiser & Sophie
Tooth wear as a means to quantify intra-specific variations in diet and chewing movements - Scientific Reports 2016, 6:3037 Ivan Calandra, Gaëlle Labonne, Ellen Schulz-Kornas, Thomas M. Kaiser & Sophie
State of the Union... Functional Genomics Research Stream. Molecular Biology. Genomics. Computational Biology
 Functional Genomics Research Stream State of the Union... Research Meeting: February 16, 2010 Functional Genomics & Research Report III Concepts Genomics Molecular Biology Computational Biology Genome
Functional Genomics Research Stream State of the Union... Research Meeting: February 16, 2010 Functional Genomics & Research Report III Concepts Genomics Molecular Biology Computational Biology Genome
Efficiency change over time
 Efficiency change over time Heikki Tikanmäki Optimointiopin seminaari 14.11.2007 Contents Introduction (11.1) Window analysis (11.2) Example, application, analysis Malmquist index (11.3) Dealing with panel
Efficiency change over time Heikki Tikanmäki Optimointiopin seminaari 14.11.2007 Contents Introduction (11.1) Window analysis (11.2) Example, application, analysis Malmquist index (11.3) Dealing with panel
Other approaches to restrict multipliers
 Other approaches to restrict multipliers Heikki Tikanmäki Optimointiopin seminaari 10.10.2007 Contents Short revision (6.2) Another Assurance Region Model (6.3) Cone-Ratio Method (6.4) An Application of
Other approaches to restrict multipliers Heikki Tikanmäki Optimointiopin seminaari 10.10.2007 Contents Short revision (6.2) Another Assurance Region Model (6.3) Cone-Ratio Method (6.4) An Application of
Functional Genomics & Proteomics
 Functional Genomics & Proteomics Genome Sequences TCACAATTTAGACATCTAGTCTTCCACTTAAGCATATTTAGATTGTTTCCAGTTTTCAGCTTTTATGACTAAATCTTCTAAAATTGTTTTTCCCTAAATGTATATTTTAATTTGTCTCAGGAGTAGAATTTCTGAGTCATAAAGCGGT CATATGTATAAATTTTAGGTGCCTCATAGCTCTTCAAATAGTCATCCCATTTTATACATCCAGGCAATATATGAGAGTTCTTGGTGCTCCACATCTTAGCTAGGATTTGATGTCAACCAGTCTCTTTAATTTAGATATTCTAGTACAT
Functional Genomics & Proteomics Genome Sequences TCACAATTTAGACATCTAGTCTTCCACTTAAGCATATTTAGATTGTTTCCAGTTTTCAGCTTTTATGACTAAATCTTCTAAAATTGTTTTTCCCTAAATGTATATTTTAATTTGTCTCAGGAGTAGAATTTCTGAGTCATAAAGCGGT CATATGTATAAATTTTAGGTGCCTCATAGCTCTTCAAATAGTCATCCCATTTTATACATCCAGGCAATATATGAGAGTTCTTGGTGCTCCACATCTTAGCTAGGATTTGATGTCAACCAGTCTCTTTAATTTAGATATTCTAGTACAT
Gap-filling methods for CH 4 data
 Gap-filling methods for CH 4 data Sigrid Dengel University of Helsinki Outline - Ecosystems known for CH 4 emissions; - Why is gap-filling of CH 4 data not as easy and straight forward as CO 2 ; - Gap-filling
Gap-filling methods for CH 4 data Sigrid Dengel University of Helsinki Outline - Ecosystems known for CH 4 emissions; - Why is gap-filling of CH 4 data not as easy and straight forward as CO 2 ; - Gap-filling
Alternative DEA Models
 Mat-2.4142 Alternative DEA Models 19.9.2007 Table of Contents Banker-Charnes-Cooper Model Additive Model Example Data Home assignment BCC Model (Banker-Charnes-Cooper) production frontiers spanned by convex
Mat-2.4142 Alternative DEA Models 19.9.2007 Table of Contents Banker-Charnes-Cooper Model Additive Model Example Data Home assignment BCC Model (Banker-Charnes-Cooper) production frontiers spanned by convex
16. Allocation Models
 16. Allocation Models Juha Saloheimo 17.1.27 S steemianalsin Optimointiopin seminaari - Sks 27 Content Introduction Overall Efficienc with common prices and costs Cost Efficienc S steemianalsin Revenue
16. Allocation Models Juha Saloheimo 17.1.27 S steemianalsin Optimointiopin seminaari - Sks 27 Content Introduction Overall Efficienc with common prices and costs Cost Efficienc S steemianalsin Revenue
Bounds on non-surjective cellular automata
 Bounds on non-surjective cellular automata Jarkko Kari Pascal Vanier Thomas Zeume University of Turku LIF Marseille Universität Hannover 27 august 2009 J. Kari, P. Vanier, T. Zeume (UTU) Bounds on non-surjective
Bounds on non-surjective cellular automata Jarkko Kari Pascal Vanier Thomas Zeume University of Turku LIF Marseille Universität Hannover 27 august 2009 J. Kari, P. Vanier, T. Zeume (UTU) Bounds on non-surjective
voidaan hylätä, pienempi vai suurempi kuin 1 %?
 [TILTP1] TILASTOTIETEEN JOHDANTOKURSSI, Syksy 2011 http://www.uta.fi/~strale/tiltp1/index.html 30.9.2011 klo 13:07:54 HARJOITUS 5 viikko 41 Ryhmät ke 08.30 10.00 ls. C8 Leppälä to 12.15 13.45 ls. A2a Laine
[TILTP1] TILASTOTIETEEN JOHDANTOKURSSI, Syksy 2011 http://www.uta.fi/~strale/tiltp1/index.html 30.9.2011 klo 13:07:54 HARJOITUS 5 viikko 41 Ryhmät ke 08.30 10.00 ls. C8 Leppälä to 12.15 13.45 ls. A2a Laine
1. SIT. The handler and dog stop with the dog sitting at heel. When the dog is sitting, the handler cues the dog to heel forward.
 START START SIT 1. SIT. The handler and dog stop with the dog sitting at heel. When the dog is sitting, the handler cues the dog to heel forward. This is a static exercise. SIT STAND 2. SIT STAND. The
START START SIT 1. SIT. The handler and dog stop with the dog sitting at heel. When the dog is sitting, the handler cues the dog to heel forward. This is a static exercise. SIT STAND 2. SIT STAND. The
Plasmid construction PCR reactions were carried out with Pfu or Pfu turbo polymerases (Stratagene) unless otherwise stated.
 Expanding the Genetic Code of Yeast for Incorporation of Diverse Unnatural Amino Acids via a Pyrrolysyl-tRNA Synthetase/tRNA Pair Susan M. Hancock 1, Rajendra Uprety 2, Alexander Deiters 2 & Jason W. Chin
Expanding the Genetic Code of Yeast for Incorporation of Diverse Unnatural Amino Acids via a Pyrrolysyl-tRNA Synthetase/tRNA Pair Susan M. Hancock 1, Rajendra Uprety 2, Alexander Deiters 2 & Jason W. Chin
Online Supplement 1 Foudi et al. 2008
 Online Supplement 1 Foudi et al. 2008 This supplementary file contains: I) Supplementary Figures 1-10 II) Supplementary References (38-42) M2-rtT ROS26 S M2-rtT -globin poly + doxycycline S+poly TetOP
Online Supplement 1 Foudi et al. 2008 This supplementary file contains: I) Supplementary Figures 1-10 II) Supplementary References (38-42) M2-rtT ROS26 S M2-rtT -globin poly + doxycycline S+poly TetOP
Kysymys 5 Compared to the workload, the number of credits awarded was (1 credits equals 27 working hours): (4)
 Tilasto T1106120-s2012palaute Kyselyn T1106120+T1106120-s2012palaute yhteenveto: vastauksia (4) Kysymys 1 Degree programme: (4) TIK: TIK 1 25% ************** INF: INF 0 0% EST: EST 0 0% TLT: TLT 0 0% BIO:
Tilasto T1106120-s2012palaute Kyselyn T1106120+T1106120-s2012palaute yhteenveto: vastauksia (4) Kysymys 1 Degree programme: (4) TIK: TIK 1 25% ************** INF: INF 0 0% EST: EST 0 0% TLT: TLT 0 0% BIO:
Tässä harjoituksessa käydään läpi R-ohjelman käyttöä esimerkkidatan avulla. eli matriisissa on 200 riviä (havainnot) ja 7 saraketta (mittaus-arvot)
 R-ohjelman käyttö data-analyysissä Panu Somervuo 2014 Tässä harjoituksessa käydään läpi R-ohjelman käyttöä esimerkkidatan avulla. 0) käynnistetään R-ohjelma Huom.1 allaolevissa ohjeissa '>' merkki on R:n
R-ohjelman käyttö data-analyysissä Panu Somervuo 2014 Tässä harjoituksessa käydään läpi R-ohjelman käyttöä esimerkkidatan avulla. 0) käynnistetään R-ohjelma Huom.1 allaolevissa ohjeissa '>' merkki on R:n
Capacity Utilization
 Capacity Utilization Tim Schöneberg 28th November Agenda Introduction Fixed and variable input ressources Technical capacity utilization Price based capacity utilization measure Long run and short run
Capacity Utilization Tim Schöneberg 28th November Agenda Introduction Fixed and variable input ressources Technical capacity utilization Price based capacity utilization measure Long run and short run
Eukaryotic Comparative Genomics
 Eukaryotic Comparative Genomics Detecting Conserved Sequences Charles Darwin Motoo Kimura Evolution of Neutral DNA A A T C TA AT T G CT G T GA T T C A GA G T A G CA G T GA AT A GT C T T T GA T GT T G T
Eukaryotic Comparative Genomics Detecting Conserved Sequences Charles Darwin Motoo Kimura Evolution of Neutral DNA A A T C TA AT T G CT G T GA T T C A GA G T A G CA G T GA AT A GT C T T T GA T GT T G T
Perhevapaiden palkkavaikutukset
 Perhevapaiden palkkavaikutukset Perhe ja ura tasa-arvon haasteena seminaari, Helsinki 20.11.2007 Jenni Kellokumpu Esityksen runko 1. Tutkimuksen tavoite 2. Teoria 3. Aineisto, tutkimusasetelma ja otos
Perhevapaiden palkkavaikutukset Perhe ja ura tasa-arvon haasteena seminaari, Helsinki 20.11.2007 Jenni Kellokumpu Esityksen runko 1. Tutkimuksen tavoite 2. Teoria 3. Aineisto, tutkimusasetelma ja otos
Supplementary Material : Baker et al. 1
 Supplementary Material : Baker et al. 1 Supplementary Material Title: Species identity and human consumption of beaked whales in the Gilbert Islands, Republic of Kiribati Authors: C. Scott Baker, Al Hutt,
Supplementary Material : Baker et al. 1 Supplementary Material Title: Species identity and human consumption of beaked whales in the Gilbert Islands, Republic of Kiribati Authors: C. Scott Baker, Al Hutt,
Green Rock Ltd Johdintie 5, FI 90630 OULU, FINLAND Tel. +358 8 8192 200 info@greenrock.fi
 Table contains test data of GR IISI S 6 unit. Tests were carried out mainly in wintertime 2009. Reduction is calculated between inlet value and outlet value of the results. IN ( ) OUT () BOD N P BOD N
Table contains test data of GR IISI S 6 unit. Tests were carried out mainly in wintertime 2009. Reduction is calculated between inlet value and outlet value of the results. IN ( ) OUT () BOD N P BOD N
MTTTP5, luento Luottamusväli, määritelmä
 23.11.2017/1 MTTTP5, luento 23.11.2017 Luottamusväli, määritelmä Olkoot A ja B satunnaisotoksen perusteella määriteltyjä satunnaismuuttujia. Väli (A, B) on parametrin 100(1 - ) %:n luottamusväli, jos P(A
23.11.2017/1 MTTTP5, luento 23.11.2017 Luottamusväli, määritelmä Olkoot A ja B satunnaisotoksen perusteella määriteltyjä satunnaismuuttujia. Väli (A, B) on parametrin 100(1 - ) %:n luottamusväli, jos P(A
enzymatic families regulated in response to a short-term cytokinin treatment in root apices
 A B C D Supplemental tlfigure 1. 1Mtbli Metabolic processes, hormonal pathways and dlarge enzymatic families regulated in response to a short-term cytokinin treatment in root apices The 297 transcripts
A B C D Supplemental tlfigure 1. 1Mtbli Metabolic processes, hormonal pathways and dlarge enzymatic families regulated in response to a short-term cytokinin treatment in root apices The 297 transcripts
VIIKKI BIOCENTER University of Helsinki
 VIIKKI BIOCENTER University of Helsinki Biologian DNA koodi ja sen selvittäminen Petri Auvinen DNA Sequencing and Genomics Laboratory Institute of Biotechnology Kuinka solut kehittyivät? Kolmenlaisia soluja
VIIKKI BIOCENTER University of Helsinki Biologian DNA koodi ja sen selvittäminen Petri Auvinen DNA Sequencing and Genomics Laboratory Institute of Biotechnology Kuinka solut kehittyivät? Kolmenlaisia soluja
Supplementary Information
 Supplementary Information Supplementary Table 1. Strain table Strain name Genotype Reference CEN.PK111-27B MATa leu2 trp1 Euroscarf TC-49 TC-50 pdc5::aro4* aro10::aro7* This study TC-50 MATa leu2 trp1
Supplementary Information Supplementary Table 1. Strain table Strain name Genotype Reference CEN.PK111-27B MATa leu2 trp1 Euroscarf TC-49 TC-50 pdc5::aro4* aro10::aro7* This study TC-50 MATa leu2 trp1
3 9-VUOTIAIDEN LASTEN SUORIUTUMINEN BOSTONIN NIMENTÄTESTISTÄ
 Puhe ja kieli, 27:4, 141 147 (2007) 3 9-VUOTIAIDEN LASTEN SUORIUTUMINEN BOSTONIN NIMENTÄTESTISTÄ Soile Loukusa, Oulun yliopisto, suomen kielen, informaatiotutkimuksen ja logopedian laitos & University
Puhe ja kieli, 27:4, 141 147 (2007) 3 9-VUOTIAIDEN LASTEN SUORIUTUMINEN BOSTONIN NIMENTÄTESTISTÄ Soile Loukusa, Oulun yliopisto, suomen kielen, informaatiotutkimuksen ja logopedian laitos & University
Paikkatiedon semanttinen mallinnus, integrointi ja julkaiseminen Case Suomalainen ajallinen paikkaontologia SAPO
 Paikkatiedon semanttinen mallinnus, integrointi ja julkaiseminen Case Suomalainen ajallinen paikkaontologia SAPO Tomi Kauppinen, Eero Hyvönen, Jari Väätäinen Semantic Computing Research Group (SeCo) http://www.seco.tkk.fi/
Paikkatiedon semanttinen mallinnus, integrointi ja julkaiseminen Case Suomalainen ajallinen paikkaontologia SAPO Tomi Kauppinen, Eero Hyvönen, Jari Väätäinen Semantic Computing Research Group (SeCo) http://www.seco.tkk.fi/
FinFamily PostgreSQL installation ( ) FinFamily PostgreSQL
 FinFamily PostgreSQL 1 Sisällys / Contents FinFamily PostgreSQL... 1 1. Asenna PostgreSQL tietokanta / Install PostgreSQL database... 3 1.1. PostgreSQL tietokannasta / About the PostgreSQL database...
FinFamily PostgreSQL 1 Sisällys / Contents FinFamily PostgreSQL... 1 1. Asenna PostgreSQL tietokanta / Install PostgreSQL database... 3 1.1. PostgreSQL tietokannasta / About the PostgreSQL database...
The CCR Model and Production Correspondence
 The CCR Model and Production Correspondence Tim Schöneberg The 19th of September Agenda Introduction Definitions Production Possiblity Set CCR Model and the Dual Problem Input excesses and output shortfalls
The CCR Model and Production Correspondence Tim Schöneberg The 19th of September Agenda Introduction Definitions Production Possiblity Set CCR Model and the Dual Problem Input excesses and output shortfalls
Use of spatial data in the new production environment and in a data warehouse
 Use of spatial data in the new production environment and in a data warehouse Nordic Forum for Geostatistics 2007 Session 3, GI infrastructure and use of spatial database Statistics Finland, Population
Use of spatial data in the new production environment and in a data warehouse Nordic Forum for Geostatistics 2007 Session 3, GI infrastructure and use of spatial database Statistics Finland, Population
ECONOMIC IMPORTANCE OF THE FOREST SECTOR IN FINNISH REGIONS AND SUB-REGIONS IN 2002
 ECONOMIC IMPORTANCE OF THE FOREST SECTOR IN FINNISH REGIONS AND SUB-REGIONS IN 2002 Sources: Statistic Finland, Genimap Oy (L6022/05) The Economic Importance of the Forest Sector in Finnish Regions and
ECONOMIC IMPORTANCE OF THE FOREST SECTOR IN FINNISH REGIONS AND SUB-REGIONS IN 2002 Sources: Statistic Finland, Genimap Oy (L6022/05) The Economic Importance of the Forest Sector in Finnish Regions and
Constructive Alignment in Specialisation Studies in Industrial Pharmacy in Finland
 Constructive Alignment in Specialisation Studies in Industrial Pharmacy in Finland Anne Mari Juppo, Nina Katajavuori University of Helsinki Faculty of Pharmacy 23.7.2012 1 Background Pedagogic research
Constructive Alignment in Specialisation Studies in Industrial Pharmacy in Finland Anne Mari Juppo, Nina Katajavuori University of Helsinki Faculty of Pharmacy 23.7.2012 1 Background Pedagogic research
S-55.1100 SÄHKÖTEKNIIKKA JA ELEKTRONIIKKA
 S-55.00 SÄHKÖKNKKA A KONKKA. välikoe 2..2008. Saat vastata vain neljään tehtävään!. aske jännite U. = 4 Ω, 2 = Ω, = Ω, = 2, 2 =, = A, 2 = U 2 2 2 2. ännitelähde tuottaa hetkestä t = t < 0 alkaen kaksiportaisen
S-55.00 SÄHKÖKNKKA A KONKKA. välikoe 2..2008. Saat vastata vain neljään tehtävään!. aske jännite U. = 4 Ω, 2 = Ω, = Ω, = 2, 2 =, = A, 2 = U 2 2 2 2. ännitelähde tuottaa hetkestä t = t < 0 alkaen kaksiportaisen
Statistical design. Tuomas Selander
 Statistical design Tuomas Selander 28.8.2014 Introduction Biostatistician Work area KYS-erva KYS, Jyväskylä, Joensuu, Mikkeli, Savonlinna Work tasks Statistical methods, selection and quiding Data analysis
Statistical design Tuomas Selander 28.8.2014 Introduction Biostatistician Work area KYS-erva KYS, Jyväskylä, Joensuu, Mikkeli, Savonlinna Work tasks Statistical methods, selection and quiding Data analysis
Graph. COMPUTE x=rv.normal(0,0.04). COMPUTE y=rv.normal(0,0.04). execute.
 COMPUTE x=rv.ormal(0,0.04). COMPUTE y=rv.ormal(0,0.04). execute. compute hplib_man_r = hplib_man + x. compute arvokons_man_r = arvokons_man + y. GRAPH /SCATTERPLOT(BIVAR)=hplib_man_r WITH arvokons_man_r
COMPUTE x=rv.ormal(0,0.04). COMPUTE y=rv.ormal(0,0.04). execute. compute hplib_man_r = hplib_man + x. compute arvokons_man_r = arvokons_man + y. GRAPH /SCATTERPLOT(BIVAR)=hplib_man_r WITH arvokons_man_r
HARJOITUS- PAKETTI A
 Logistiikka A35A00310 Tuotantotalouden perusteet HARJOITUS- PAKETTI A (6 pistettä) TUTA 19 Luento 3.Ennustaminen County General 1 piste The number of heart surgeries performed at County General Hospital
Logistiikka A35A00310 Tuotantotalouden perusteet HARJOITUS- PAKETTI A (6 pistettä) TUTA 19 Luento 3.Ennustaminen County General 1 piste The number of heart surgeries performed at County General Hospital
Underpinning Phage Arbitrium Communication Systems
 Molecular Cell, Volume 74 Supplemental Information Deciphering the Molecular Mechanism Underpinning Phage Arbitrium Communication Systems Francisca Gallego del Sol, José R. Penadés, and Alberto Marina
Molecular Cell, Volume 74 Supplemental Information Deciphering the Molecular Mechanism Underpinning Phage Arbitrium Communication Systems Francisca Gallego del Sol, José R. Penadés, and Alberto Marina
Tree map system in harvester
 Tree map system in harvester Fibic seminar 12.6.2013 Lahti Timo Melkas, Metsäteho Oy Mikko Miettinen, Argone Oy Kalle Einola, Ponsse Oyj Project goals EffFibre project 2011-2013 (WP3) To evaluate the accuracy
Tree map system in harvester Fibic seminar 12.6.2013 Lahti Timo Melkas, Metsäteho Oy Mikko Miettinen, Argone Oy Kalle Einola, Ponsse Oyj Project goals EffFibre project 2011-2013 (WP3) To evaluate the accuracy
Accommodation statistics
 Transport and Tourism 2011 Accommodation statistics 2011, January Nights spent by foreign tourists in Finland increased by per cent in January The number of recorded nights spent by foreign tourists at
Transport and Tourism 2011 Accommodation statistics 2011, January Nights spent by foreign tourists in Finland increased by per cent in January The number of recorded nights spent by foreign tourists at
Lisätehtäviä ratkaisuineen luentomonisteen lukuun 6 liittyen., jos otoskeskiarvo on suurempi kuin 13,96. Mikä on testissä käytetty α:n arvo?
 MTTTP5, kevät 2016 15.2.2016/RL Lisätehtäviä ratkaisuineen luentomonisteen lukuun 6 liittyen 1. Valitaan 25 alkion satunnaisotos jakaumasta N(µ, 25). Olkoon H 0 : µ = 12. Hylätään H 0, jos otoskeskiarvo
MTTTP5, kevät 2016 15.2.2016/RL Lisätehtäviä ratkaisuineen luentomonisteen lukuun 6 liittyen 1. Valitaan 25 alkion satunnaisotos jakaumasta N(µ, 25). Olkoon H 0 : µ = 12. Hylätään H 0, jos otoskeskiarvo
Tavaroiden ulkomaankauppatilastojen tulkinnan haasteet. 22.3.2012 Timo Koskimäki
 Tavaroiden ulkomaankauppatilastojen tulkinnan haasteet 22.3.2012 Timo Koskimäki 1 Sisältö Johdannoksi Esimerkit Mikro: Kännykän arvonlisän komponentit Makro: Suomen kauppatase ja viestintäklusteri Kauppatilastojen
Tavaroiden ulkomaankauppatilastojen tulkinnan haasteet 22.3.2012 Timo Koskimäki 1 Sisältö Johdannoksi Esimerkit Mikro: Kännykän arvonlisän komponentit Makro: Suomen kauppatase ja viestintäklusteri Kauppatilastojen
Vaisala s New Global L ightning Lightning Dataset GLD360
 Vaisala s New Global Lightning Dataset GLD360 Vaisala Global Lightning Dataset GLD360 Page 2 / Oct09 / Holle-SW Hydro / Vaisala Schedule GLD360 Validation Applications Demonstration Page 3 / Oct09 / Holle-SW
Vaisala s New Global Lightning Dataset GLD360 Vaisala Global Lightning Dataset GLD360 Page 2 / Oct09 / Holle-SW Hydro / Vaisala Schedule GLD360 Validation Applications Demonstration Page 3 / Oct09 / Holle-SW
Mobility Tool. Demo 21.8.2013 CIMO
 Mobility Tool Demo 21.8.2013 CIMO 1 Sisältö Korkeakoulukäyttäjä (Beneficiary User) Home Project details Partners Mobilities Working with CSV Import Mobilities Budget 2 Home Find your project Project menu
Mobility Tool Demo 21.8.2013 CIMO 1 Sisältö Korkeakoulukäyttäjä (Beneficiary User) Home Project details Partners Mobilities Working with CSV Import Mobilities Budget 2 Home Find your project Project menu
Efficient regeneration system and Agrobacteriummediated transformation of Vetiver. PT 1
 Efficient regeneration system and Agrobacteriummediated transformation of Vetiver PT www.acolor.net 1 Outline: Part I: Why (Background) Part II: Some concrete work we have done Part III: Prospects to further
Efficient regeneration system and Agrobacteriummediated transformation of Vetiver PT www.acolor.net 1 Outline: Part I: Why (Background) Part II: Some concrete work we have done Part III: Prospects to further
Vakiomallisten kuumakanavajärjestelmien kytkeminen Wiring of standard hotrunner systems
 Teknillinen dokumentaatio Technical documentation E. Vakiomallisten kuumakanavajärjestelmien kytkeminen Wiring of standard hotrunner systems Version.0 ! Tärkeä turvaohje / Important safety instruction
Teknillinen dokumentaatio Technical documentation E. Vakiomallisten kuumakanavajärjestelmien kytkeminen Wiring of standard hotrunner systems Version.0 ! Tärkeä turvaohje / Important safety instruction
Perusnäkymä yksisuuntaiseen ANOVAaan
 Metsämuuronen 2006. TTP Tutkimuksen tekemisen perusteet ihmistieteissä Taulukko.51.1 Analyysiin mukaan tulevat muuttujat Mja selite Merkitys mallissa F1 Ensimmäinen faktoripistemuuttuja Selitettävä muuttuja
Metsämuuronen 2006. TTP Tutkimuksen tekemisen perusteet ihmistieteissä Taulukko.51.1 Analyysiin mukaan tulevat muuttujat Mja selite Merkitys mallissa F1 Ensimmäinen faktoripistemuuttuja Selitettävä muuttuja
OHJ-7600 Ihminen ja tekniikka -seminaari, 4 op Käyttäjäkokemuksen kvantitatiivinen analyysi. Luento 3
 OHJ-7600 Ihminen ja tekniikka -seminaari, 4 op Käyttäjäkokemuksen kvantitatiivinen analyysi Luento 3 Tutkimussuunnitelman rakenne-ehdotus Otsikko 1. Motivaatio/tausta 2. Tutkimusaihe/ -tavoitteet ja kysymykset
OHJ-7600 Ihminen ja tekniikka -seminaari, 4 op Käyttäjäkokemuksen kvantitatiivinen analyysi Luento 3 Tutkimussuunnitelman rakenne-ehdotus Otsikko 1. Motivaatio/tausta 2. Tutkimusaihe/ -tavoitteet ja kysymykset
Pricing policy: The Finnish experience
 Pricing policy: The Finnish experience Esa Österberg Senior Researcher Alcohol and Drug Research, STAKES, Helsinki, Finland esa.osterberg@stakes.fi Three pillars of traditional Nordic alcohol control Strict
Pricing policy: The Finnish experience Esa Österberg Senior Researcher Alcohol and Drug Research, STAKES, Helsinki, Finland esa.osterberg@stakes.fi Three pillars of traditional Nordic alcohol control Strict
VIIKKI BIOCENTER University of Helsinki
 VIIKKI BIOCENTER University of Helsinki Mitä uudet DNA sekvensointimenetelmät voivat paljastaa luonnonjärjestelmästä? Petri Auvinen DNA Sequencing and Genomics Laboratory Institute of Biotechnology Petri
VIIKKI BIOCENTER University of Helsinki Mitä uudet DNA sekvensointimenetelmät voivat paljastaa luonnonjärjestelmästä? Petri Auvinen DNA Sequencing and Genomics Laboratory Institute of Biotechnology Petri
7.4 Variability management
 7.4 Variability management time... space software product-line should support variability in space (different products) support variability in time (maintenance, evolution) 1 Product variation Product
7.4 Variability management time... space software product-line should support variability in space (different products) support variability in time (maintenance, evolution) 1 Product variation Product
MCV 1,5/ 3-G-3,81. Poimi online-katalogista. Tilausnumero:
 Poimi online-katalogista MCV 1,5/ 3-G-3,81 Tilausnumero: 1803439 The figure shows a 10-position version of the product Header, Nominal current:, Nom. voltage: 160 V, Pitch: 3.81 mm, Number of positions:
Poimi online-katalogista MCV 1,5/ 3-G-3,81 Tilausnumero: 1803439 The figure shows a 10-position version of the product Header, Nominal current:, Nom. voltage: 160 V, Pitch: 3.81 mm, Number of positions:
EKOSYSTEEMIT INVEST IN TYÖKALUNA?
 EKOSYSTEEMIT INVEST IN TYÖKALUNA? Mitä Invest in -tapahtumat ovat? M&A T&K&I toimintaa GREEN FIELD Jackpot Pääomasijoitus Tutkimus -rahaa Grants Uusi tuotannollinen yritys suomeen Green Field Yritysosto
EKOSYSTEEMIT INVEST IN TYÖKALUNA? Mitä Invest in -tapahtumat ovat? M&A T&K&I toimintaa GREEN FIELD Jackpot Pääomasijoitus Tutkimus -rahaa Grants Uusi tuotannollinen yritys suomeen Green Field Yritysosto
KONEISTUSKOKOONPANON TEKEMINEN NX10-YMPÄRISTÖSSÄ
 KONEISTUSKOKOONPANON TEKEMINEN NX10-YMPÄRISTÖSSÄ https://community.plm.automation.siemens.com/t5/tech-tips- Knowledge-Base-NX/How-to-simulate-any-G-code-file-in-NX- CAM/ta-p/3340 Koneistusympäristön määrittely
KONEISTUSKOKOONPANON TEKEMINEN NX10-YMPÄRISTÖSSÄ https://community.plm.automation.siemens.com/t5/tech-tips- Knowledge-Base-NX/How-to-simulate-any-G-code-file-in-NX- CAM/ta-p/3340 Koneistusympäristön määrittely
Network to Get Work. Tehtäviä opiskelijoille Assignments for students. www.laurea.fi
 Network to Get Work Tehtäviä opiskelijoille Assignments for students www.laurea.fi Ohje henkilöstölle Instructions for Staff Seuraavassa on esitetty joukko tehtäviä, joista voit valita opiskelijaryhmällesi
Network to Get Work Tehtäviä opiskelijoille Assignments for students www.laurea.fi Ohje henkilöstölle Instructions for Staff Seuraavassa on esitetty joukko tehtäviä, joista voit valita opiskelijaryhmällesi
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
ELEMET- MOCASTRO. Effect of grain size on A 3 temperatures in C-Mn and low alloyed steels - Gleeble tests and predictions. Period
 1 ELEMET- MOCASTRO Effect of grain size on A 3 temperatures in C-Mn and low alloyed steels - Gleeble tests and predictions Period 20.02-25.05.2012 Diaarinumero Rahoituspäätöksen numero 1114/31/2010 502/10
1 ELEMET- MOCASTRO Effect of grain size on A 3 temperatures in C-Mn and low alloyed steels - Gleeble tests and predictions Period 20.02-25.05.2012 Diaarinumero Rahoituspäätöksen numero 1114/31/2010 502/10
Metsälamminkankaan tuulivoimapuiston osayleiskaava
 VAALAN KUNTA TUULISAIMAA OY Metsälamminkankaan tuulivoimapuiston osayleiskaava Liite 3. Varjostusmallinnus FCG SUUNNITTELU JA TEKNIIKKA OY 12.5.2015 P25370 SHADOW - Main Result Assumptions for shadow calculations
VAALAN KUNTA TUULISAIMAA OY Metsälamminkankaan tuulivoimapuiston osayleiskaava Liite 3. Varjostusmallinnus FCG SUUNNITTELU JA TEKNIIKKA OY 12.5.2015 P25370 SHADOW - Main Result Assumptions for shadow calculations
A DEA Game II. Juha Saloheimo S ysteemianalyysin. Laboratorio. Teknillinen korkeakoulu
 A DEA Game II Juha Salohemo 12.12.2007 Content Recap of the Example The Shapley Value Margnal Contrbuton, Ordered Coaltons, Soluton to the Example DEA Mn Game Summary Home Assgnment Recap of the Example
A DEA Game II Juha Salohemo 12.12.2007 Content Recap of the Example The Shapley Value Margnal Contrbuton, Ordered Coaltons, Soluton to the Example DEA Mn Game Summary Home Assgnment Recap of the Example
Somaattinen sairaus nuoruudessa ja mielenterveyden häiriön puhkeamisen riski
 + Somaattinen sairaus nuoruudessa ja mielenterveyden häiriön puhkeamisen riski LINNEA KARLSSON + Riskitekijöitä n Ulkonäköön liittyvät muutokset n Toimintakyvyn menetykset n Ikätovereista eroon joutuminen
+ Somaattinen sairaus nuoruudessa ja mielenterveyden häiriön puhkeamisen riski LINNEA KARLSSON + Riskitekijöitä n Ulkonäköön liittyvät muutokset n Toimintakyvyn menetykset n Ikätovereista eroon joutuminen
11/17/11. Gene Regulation. Gene Regulation. Gene Regulation. Finding Regulatory Motifs in DNA Sequences. Regulatory Proteins
 Gene Regulation Finding Regulatory Motifs in DNA Sequences An experiment shows that when X is knocked out, 20 other s are not expressed How can one have such drastic effects? Regulatory Proteins Gene X
Gene Regulation Finding Regulatory Motifs in DNA Sequences An experiment shows that when X is knocked out, 20 other s are not expressed How can one have such drastic effects? Regulatory Proteins Gene X
S Sähkön jakelu ja markkinat S Electricity Distribution and Markets
 S-18.3153 Sähkön jakelu ja markkinat S-18.3154 Electricity Distribution and Markets Voltage Sag 1) Kolmivaiheinen vastukseton oikosulku tapahtuu 20 kv lähdöllä etäisyydellä 1 km, 3 km, 5 km, 8 km, 10 km
S-18.3153 Sähkön jakelu ja markkinat S-18.3154 Electricity Distribution and Markets Voltage Sag 1) Kolmivaiheinen vastukseton oikosulku tapahtuu 20 kv lähdöllä etäisyydellä 1 km, 3 km, 5 km, 8 km, 10 km
Digital Admap Native. Campaign: Kesko supermarket
 Digital Admap Native Campaign: Kesko supermarket Digital Admap Native Campaign: Kesko Supermarket Mainosmuoto: Natiivi Media: IS.fi Campaign period: 25 September Date of measurement: 26 September Unique:
Digital Admap Native Campaign: Kesko supermarket Digital Admap Native Campaign: Kesko Supermarket Mainosmuoto: Natiivi Media: IS.fi Campaign period: 25 September Date of measurement: 26 September Unique:
B3206 Microbial Genetics
 Prof. Fahd M. Nasr Lebanese university Faculty of sciences I Department of Natural Sciences fnasr@ul.edu.lb B3206 Microbial Genetics 1 Eukaryotic M. G. The yeast Saccharomyces cerevisiae as a genetic model
Prof. Fahd M. Nasr Lebanese university Faculty of sciences I Department of Natural Sciences fnasr@ul.edu.lb B3206 Microbial Genetics 1 Eukaryotic M. G. The yeast Saccharomyces cerevisiae as a genetic model
MC 1,5/10-G-3,81. Poimi online-katalogista. Tilausnumero:
 Poimi online-katalogista MC 1,5/10-G-3,81 Tilausnumero: 1803358 The figure shows a 10-position version of the product Header, Nominal current:, Nom. voltage: 160 V, Pitch: 3.81 mm, Number of positions:
Poimi online-katalogista MC 1,5/10-G-3,81 Tilausnumero: 1803358 The figure shows a 10-position version of the product Header, Nominal current:, Nom. voltage: 160 V, Pitch: 3.81 mm, Number of positions:
Telecommunication Software
 Telecommunication Software Final exam 21.11.2006 COMPUTER ENGINEERING LABORATORY 521265A Vastaukset englanniksi tai suomeksi. / Answers in English or in Finnish. 1. (a) Määrittele sovellusviesti, PersonnelRecord,
Telecommunication Software Final exam 21.11.2006 COMPUTER ENGINEERING LABORATORY 521265A Vastaukset englanniksi tai suomeksi. / Answers in English or in Finnish. 1. (a) Määrittele sovellusviesti, PersonnelRecord,
Research plan for masters thesis in forest sciences. The PELLETime 2009 Symposium Mervi Juntunen
 Modelling tree and stand characteristics and estimating biomass removals and harvesting costs of lodgepole pine (Pinus contorta) plantations in Iceland Research plan for masters thesis in forest sciences
Modelling tree and stand characteristics and estimating biomass removals and harvesting costs of lodgepole pine (Pinus contorta) plantations in Iceland Research plan for masters thesis in forest sciences
 Digitally signed by Hans Vadbäck DN: cn=hans Vadbäck, o, ou=fcg Suunnittelu ja Tekniikka Oy, email=hans.vadback@fcg.fi, c=fi Date: 2016.12.20 15:45:35 +02'00' Jakob Kjellman Digitally signed by Jakob Kjellman
Digitally signed by Hans Vadbäck DN: cn=hans Vadbäck, o, ou=fcg Suunnittelu ja Tekniikka Oy, email=hans.vadback@fcg.fi, c=fi Date: 2016.12.20 15:45:35 +02'00' Jakob Kjellman Digitally signed by Jakob Kjellman
Frequencies. Frequency Table
 GET FILE='C:\Documents and Settings\haukkala\My Documents\kvanti\kvanti_harjo'+ '_label.sav'. DATASET NAME DataSet WINDOW=FRONT. FREQUENCIES VARIABLES=koulv paino /ORDER= ANALYSIS. Frequencies [DataSet]
GET FILE='C:\Documents and Settings\haukkala\My Documents\kvanti\kvanti_harjo'+ '_label.sav'. DATASET NAME DataSet WINDOW=FRONT. FREQUENCIES VARIABLES=koulv paino /ORDER= ANALYSIS. Frequencies [DataSet]
Bioinformatics. Sequence Analysis: Part III. Pattern Searching and Gene Finding. Fran Lewitter, Ph.D. Head, Biocomputing Whitehead Institute
 Bioinformatics Sequence Analysis: Part III. Pattern Searching and Gene Finding Fran Lewitter, Ph.D. Head, Biocomputing Whitehead Institute Course Syllabus Jan 7 Jan 14 Jan 21 Jan 28 Feb 4 Feb 11 Feb 18
Bioinformatics Sequence Analysis: Part III. Pattern Searching and Gene Finding Fran Lewitter, Ph.D. Head, Biocomputing Whitehead Institute Course Syllabus Jan 7 Jan 14 Jan 21 Jan 28 Feb 4 Feb 11 Feb 18
WindPRO version joulu 2012 Printed/Page :42 / 1. SHADOW - Main Result
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table 13.6.2013 19:42 / 1 Minimum
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table 13.6.2013 19:42 / 1 Minimum
Python Libraries 1 / 14
 Python Libraries 1 / 14 Gzip, CSV https://docs.python.org/3/library/gzip.html https://docs.python.org/3/library/csv.html https://daler.github.io/pybedtools/ - PyBed tools http://biopython.org/wiki/gff_parsing
Python Libraries 1 / 14 Gzip, CSV https://docs.python.org/3/library/gzip.html https://docs.python.org/3/library/csv.html https://daler.github.io/pybedtools/ - PyBed tools http://biopython.org/wiki/gff_parsing
( ( OX2 Perkkiö. Rakennuskanta. Varjostus. 9 x N131 x HH145
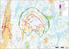 OX2 9 x N131 x HH145 Rakennuskanta Asuinrakennus Lomarakennus Liike- tai julkinen rakennus Teollinen rakennus Kirkko tai kirkollinen rak. Muu rakennus Allas Varjostus 1 h/a 8 h/a 20 h/a 0 0,5 1 1,5 2 km
OX2 9 x N131 x HH145 Rakennuskanta Asuinrakennus Lomarakennus Liike- tai julkinen rakennus Teollinen rakennus Kirkko tai kirkollinen rak. Muu rakennus Allas Varjostus 1 h/a 8 h/a 20 h/a 0 0,5 1 1,5 2 km
Tynnyrivaara, OX2 Tuulivoimahanke. ( Layout 9 x N131 x HH145. Rakennukset Asuinrakennus Lomarakennus 9 x N131 x HH145 Varjostus 1 h/a 8 h/a 20 h/a
 , Tuulivoimahanke Layout 9 x N131 x HH145 Rakennukset Asuinrakennus Lomarakennus 9 x N131 x HH145 Varjostus 1 h/a 8 h/a 20 h/a 0 0,5 1 1,5 km 2 SHADOW - Main Result Assumptions for shadow calculations
, Tuulivoimahanke Layout 9 x N131 x HH145 Rakennukset Asuinrakennus Lomarakennus 9 x N131 x HH145 Varjostus 1 h/a 8 h/a 20 h/a 0 0,5 1 1,5 km 2 SHADOW - Main Result Assumptions for shadow calculations
Asiakaspalautteen merkitys laboratoriovirheiden paljastamisessa. Taustaa
 Asiakaspalautteen merkitys laboratoriovirheiden paljastamisessa Paula Oja, TtT Laboratorio, Oulun yliopistollinen sairaala Potilasturvallisuustutkimuksen päivät 26. 27.1.2011 1 Taustaa Laboratorion tulee
Asiakaspalautteen merkitys laboratoriovirheiden paljastamisessa Paula Oja, TtT Laboratorio, Oulun yliopistollinen sairaala Potilasturvallisuustutkimuksen päivät 26. 27.1.2011 1 Taustaa Laboratorion tulee
Supporting Information Table S1 Core regions of 18 identified promoters in C. acetobutylicum
 Supporting Information Table S1 Core regions of 18 identified promoters in C. acetobutylicum Gene Core regions of the promoters Reference 35 region 10 region thl TATA TTGATA AAAATAATAATAGTGGG TATAAT TAA
Supporting Information Table S1 Core regions of 18 identified promoters in C. acetobutylicum Gene Core regions of the promoters Reference 35 region 10 region thl TATA TTGATA AAAATAATAATAGTGGG TATAAT TAA
Chapter 7. Motif finding (week 11) Chapter 8. Sequence binning (week 11)
 Course organization Introduction ( Week 1) Part I: Algorithms for Sequence Analysis (Week 1-11) Chapter 1-3, Models and theories» Probability theory and Statistics (Week 2)» Algorithm complexity analysis
Course organization Introduction ( Week 1) Part I: Algorithms for Sequence Analysis (Week 1-11) Chapter 1-3, Models and theories» Probability theory and Statistics (Week 2)» Algorithm complexity analysis
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
Eija Lahtinen Uudet kelikamerat Kaakkois-Suomen tiepiiri
 Eija Lahtinen Uudet kelikamerat Kaakkois-Suomen tiepiiri VIKING Eija Lahtinen Uudet kelikamerat Kaakkois-Suomen tiepiiri Tiehallinto Kaakkois-Suomen tiepiiri Liikenteen palvelut Kouvola 2001 Raportin
Eija Lahtinen Uudet kelikamerat Kaakkois-Suomen tiepiiri VIKING Eija Lahtinen Uudet kelikamerat Kaakkois-Suomen tiepiiri Tiehallinto Kaakkois-Suomen tiepiiri Liikenteen palvelut Kouvola 2001 Raportin
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.9.269
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.9.269
lpar1 IPB004065, IPB002277, and IPB Restriction Enyzme Differences from REBASE Gained in Variant Lost from Reference
 lpar1 IPB465, IPB2277, and IPB5385 Genomic Sequence Coding Sequence For help interpreting these results, view the PARSENP Introduction page. # View On Sequence Nucleotide Change Effect Restriction Enyzme
lpar1 IPB465, IPB2277, and IPB5385 Genomic Sequence Coding Sequence For help interpreting these results, view the PARSENP Introduction page. # View On Sequence Nucleotide Change Effect Restriction Enyzme
Vallila coated fabrics. Vallila vahakankaat
 Vallila coated fabrics Vallila vahakankaat Acrylic coated fabrics (100% cotton) 2 Africa 120405 Design: Howard Smith Aurajoki 001765 Design: Riina Kuikka 122 blue 8 beige Artisokka 001680 Design: Saara
Vallila coated fabrics Vallila vahakankaat Acrylic coated fabrics (100% cotton) 2 Africa 120405 Design: Howard Smith Aurajoki 001765 Design: Riina Kuikka 122 blue 8 beige Artisokka 001680 Design: Saara
Supplementary Table S1. Genes that are upregulated during iron toxicity. Foldchange
 Supplementary Table S1. Genes that are upregulated during iron toxicity. Locus tag Spy Number a Foldchange b Annotation c Iron efflux pmta SpyM3_1093 17.6 Iron efflux pump Arginine and serine catabolism
Supplementary Table S1. Genes that are upregulated during iron toxicity. Locus tag Spy Number a Foldchange b Annotation c Iron efflux pmta SpyM3_1093 17.6 Iron efflux pump Arginine and serine catabolism
The relationship between leisuretime physical activity and work stress with special reference to heart rate variability analyses
 The relationship between leisuretime physical activity and work stress with special reference to heart rate variability analyses Teisala Tiina, TtM, tohtorikoulutettava Jyväskylän yliopisto Terveystieteiden
The relationship between leisuretime physical activity and work stress with special reference to heart rate variability analyses Teisala Tiina, TtM, tohtorikoulutettava Jyväskylän yliopisto Terveystieteiden
Valuation of Asian Quanto- Basket Options
 Valuation of Asian Quanto- Basket Options (Final Presentation) 21.11.2011 Thesis Instructor and Supervisor: Prof. Ahti Salo Työn saa tallentaa ja julkistaa Aalto-yliopiston avoimilla verkkosivuilla. Muilta
Valuation of Asian Quanto- Basket Options (Final Presentation) 21.11.2011 Thesis Instructor and Supervisor: Prof. Ahti Salo Työn saa tallentaa ja julkistaa Aalto-yliopiston avoimilla verkkosivuilla. Muilta
Lab SBS3.FARM_Hyper-V - Navigating a SharePoint site
 Lab SBS3.FARM_Hyper-V - Navigating a SharePoint site Note! Before starting download and install a fresh version of OfficeProfessionalPlus_x64_en-us. The instructions are in the beginning of the exercise.
Lab SBS3.FARM_Hyper-V - Navigating a SharePoint site Note! Before starting download and install a fresh version of OfficeProfessionalPlus_x64_en-us. The instructions are in the beginning of the exercise.
Effect of tropical sea surface temperatures on meridional moisture transport in the southern hemisphere
 Effect of tropical sea surface temperatures on meridional moisture transport in the southern hemisphere Victoria Sinclair1 & Jennifer Catto2 Thanks to Glenn Carver, Oleg Stepanyuk and Juha Lento 1. University
Effect of tropical sea surface temperatures on meridional moisture transport in the southern hemisphere Victoria Sinclair1 & Jennifer Catto2 Thanks to Glenn Carver, Oleg Stepanyuk and Juha Lento 1. University
Supplementary Information
 Supplementary Information Quantifying Absolute Addressability in DNA origami with Molecular Resolution Maximilian T. Strauss, Florian Schueder, Daniel Haas, Philipp C. Nickels, and Ralf Jungmann Supplementary
Supplementary Information Quantifying Absolute Addressability in DNA origami with Molecular Resolution Maximilian T. Strauss, Florian Schueder, Daniel Haas, Philipp C. Nickels, and Ralf Jungmann Supplementary
TM ETRS-TM35FIN-ETRS89 WTG
 VE1 SHADOW - Main Result Calculation: 8 x Nordex N131 x HH145m Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please
VE1 SHADOW - Main Result Calculation: 8 x Nordex N131 x HH145m Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please
,0 Yes ,0 120, ,8
 SHADOW - Main Result Calculation: Alue 2 ( x 9 x HH120) TuuliSaimaa kaavaluonnos Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered
SHADOW - Main Result Calculation: Alue 2 ( x 9 x HH120) TuuliSaimaa kaavaluonnos Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered
