Supplemental Data. Manavski et al. (2015). Plant Cell /tpc
|
|
|
- Elisabet Jääskeläinen
- 5 vuotta sitten
- Katselukertoja:
Transkriptio
1 1
2 A Promoter Transcription start Physcomitrella patens TGTGTAATAATAAGTTTTAATACCTGCCCCAGAG-AACTTCTCTATCATAGTCGTTCCGGTTCTAAAC---TGATCGAAA AGGAGT-- Marchantia polymorpha TGTGTCATAATAAATTTGAAGACCTGCCT-AGGT-AATTTCTGTATAATAGTTGTTCCAGTTTTAAAC---TGA--GAAA GAGATCTA Psilotum nudum TGTGTCATAATAAATTTGAATACCTGCCCTAGGT-AACTTCTGTATTATAGTTGCTCCAGTTTTAAAC---TGAA-AAAA AAGATC-A Sorghum bicolor TATGTCATAATAGATCCGAACACTTGCCTTGGATTGACTTCAATAT-ATAATTGCTCCAGTGAATAAC---TAAAAAAAA---ATAGAAGGACGGTAGATAGTAAAGAAA-- Oryza sativa TATGTCATAATAGATCCGAACACTTGCCTCAGATTGACTTCAATAT-ATAATTGCTCCAGTGAATAAC---TAAAAAAAA---ATAGAAGGACGGTAGATAGTAAAGAAA-- Polupus trichocarpa TATGTCATAATAGATCCGAACACTTGCCCTGGATCGACTTCCAGATCATAATTGCTCTAGTGAATAAC---TAACGAAAC TAGATA-----GAAA-- Arabidopsis thaliana TATGTCATAATAGATCCGAACACTTGCCCCAGATCGACTTCCAGATCATAATTGCTCTAGTGAATAAC---TAAAGAAAATAGATGAATAGATGGAAGATAGAAGAGAAAGA Selaginella m. CATGCCATACTGAGATCGAGTACCCGCCCGAG-----CTCCCGCATCATAGCTGCTCCGGTCTCGGACGAATGAGTGGAA GGTGTAGTT ** *** * * ** *** * * * ** *** * ** ** ** ** * * Shine Dalgarno Start Physcomitrella patens --AGTC--TT TAAAG---ATTTCCTAGAAGTTTCCTATTAA TTGCTGGCAGGT-TGTTGCTATTCCTCGTCTCGAGAGAGGAGAATCTCAATG Marchantia polymorpha AAAGTT--ATA TATGG---ATCTCTCAGAAGTTTACTAATTA TTGTTGGTAGGT-TTTTCCTATGCCTCGTCTGAAGAGAGGAGAACCTCGATG Psilotum nudum AAAGTTGGATAATAAAATAATAAGT---ATCTTTTAGAAGTTTACTATTCA TTATTGGTGGGT-TTTTCCTATGCCTCGTCTGAAGAGAGGAGAACCTCGATG Sorghum bicolor AGGACTAATCA TAATATCTATCTTTAAAAGATTCAATAAAAA GACAGTTGGCGGGTCTCTTTGTATGTCTTGTCCGGAAAGAGGAGGA-CTTAATG Oryza sativa AGGACTAATCA TAATATCTATCTTTCAAAGGTTCACTAAAAA G----TTGGCGGGTCTCTTTGTATGTCTTGTCCGGAAAGAGGAGGA-CTTAATG Polupus trichocarpa AAAACTAATTA GAAAA---ATCTCTCATGGGTTCACTAATTACTT----GACTGTTGGCGGGTCTCTTTGTATGTGTTGTCCGGAAAGAGGAGGA-CTCAATG Arabidopsis thaliana AAAAAAAATTC TAAG---TATCTATCATCGGTTCACTAATTACTTGAGTGACTGTTGGCGGGTTTCTTTGTATGTGTTGTCCGGAAAGAGGAGGA-CTCAATG Selaginella m. GGGGGAAATTC CGAAG-----CTCACGGGAGTTTACTAATCA CTATCGGCGGGT-TTCTTCTGTGCTTCATCCGGAAAGGGGAGAACCCCGACG * ** ** * ** *** * * * * * ** * ** **** * * * * B Promoter Transcription start Arabidopsis thaliana TATGTCA TAATAGATCCGAACACTTGCCCCAGATCGACTTCCAGATCATAATTG----CTCTAG-----TGAATAACTAAAGAAAATAGATGAATAGATGGAAGAT Populus trichocarpa TATGTCA TAATAGATCCGAACACTTGCCCTGGATCGACTTCCAGATCATAATTG----CTCTAG-----TGAATAACTAACGAAACTAGAT Physcomitrella patens TGTGTCA TAATAAATTTGAATACCTGCCCTAGGTA-ACTTCTGTATTATAGTTG----CTCCAG-----TTTTAAACTGAA-AAAAAAGATCAAAAG Chlorella vulgaris AATATC TTTAAAATAAGTAAAAATAATTTGTAAACCAATAAAAAATATATTTA----TGGTAT-----AATATAACATATGATGTAAAAAAAACTA Thalassiosira p. AAATTCGCTAATTATTGTTTATTAACAGTGTAATTTAATTGTAAGACAAAAAATTGTATAAATT----TTCTTTTTGAGAAAATGAAAAATCGTGTATAATATATAA Euglena gracilis AAAACTTAAGTTCTAATTACATTTATAATTTATTTAACTTTAAGTG----AGTTGG TAGCTCAGTTGGTAGAGCACTCGG Chara vulgaris TTTG AAATCAAAAAAGAGTACAAAAT TTTTACAAATGG TCCATTAAGCGCTCTATATATATG--CCATACTACAGG Coccomyxa spec. GGAGGGGC-----CCACGCCCAAGGGCGTTCCGTCGGACGGGAAGCCCCTTTCACTAAACGGGGGGGAGGCCTTGTCCCGTGAGACATGGTAGGCCCAGACGGCGAG Shine Dalgarno Start Arabidopsis thaliana AGAAGAGAAAGAAAAAAAAATTCTAAGTATCTATCATCGGTTCACTAATTACTTGAGTGACTGTTGGCGGGTTTCTTTGTATGTGTTGTCCGG-AAAGAGGAGGAC---TCAATG Populus trichocarpa AGAAAAAAACTAATTAGAAAAATCTCTCATGGGTTCACTAATTACTT----GACTGTTGGCGGGTCTCTTTGTATGTGTTGTCCGG-AAAGAGGAGGAC---TCAATG Physcomitrell apatens TTGGATAATAAAATAATAAGTATCTTTTAGAAGTTTACTATT--CAT TATTGGTGGGTTT-TTCCTATGCCTCGTCTGA-AGAGAGGAGAACC--TCGATG Chlorella vulgaris TTGATTTTCTTAACCG-CTATTTTTTTTAAAAAT--ACTGTGGTAAG----ATTTTAAAAAAGATAATTTCGGT----AATTCAAA-TAAGGAGAATTTTTCGCCATG Thalassiosira p TGAAAACTCGTATTTAACAAATAGCTAAAAAAATTAACTATTTTTTA----TTTTTTATGAGAGTTTCATAGATTTCCGTCTCCCA-AAAGGAGAAAAT----CAATG Euglena gracilis CTTTTAACCGATCGGTCCTGGGTTCGAATCCCAGCCAACTCATTTTTA-----ATTTTGTGTAGGTGT ATGATAAT-TTAATCGAAAAT---TATATG Chara vulgaris -----TATGAAAGTCTGCTTGAATT---AATTATAGTTGTTCTAGTGATACATT----CAAAAGGAAAAAGATAGTTGTTGATTATTGACATATTTTTATTTTATATTTTGTATG Coccomyxa spec AGGGAGAGTCCCACAGGCCTGGCAGCATCTGTTGTTCTGCTGAAGGGTT----TGAGATAGGAGGGCTGAGTCTCTTTTATAGTATTTGTTTTTAGGAGAAATTCG-ATG * * *** 2
3 Supplemental Figure 2. Conservation of the psaa 5 UTR in Embryophyta. (A) Sequence alignment of the psaa 5 UTR of dicots, monocots, the fern-like vascular plant Psilotum nudum, the spikermoss Selaginella moellendorffii and mosses. The psaa promoter and conserved regions of the psaa 5 UTR in embryophyta are labeled in blue and yellow, respectively. The start codon is labeled in green. Stars indicate identical nucleotides. The bars under the alignment represent the probes psaa- 1-1 (blue), psaa-1-2 (red), and psaa-1-3 (green) used in EMSA experiments shown in Figure 7C. Accession numbers are indicated in the Method section. (B) Sequence alignment of the psaa 5 UTR of vascular plants, Physcomitrella, the diatom Thalassiosia pseudonana, Euglena gracilis, and diverse green algae. 3
4 4
5 A HCF145 SRPBCC1 (134 AA) At VWNVLTDYERLADFIPNLVWSGRIPCPHP-----GRIWLEQRGLQRALYWHIEARVVLDLHECLDSPNGRELHFSMVDGDFKKFEGKWSVKS-GIRSVGTVLSYEVNVIPRF---NFPAIFLERIIRSDLPVNLRAVARQAEK Gm VWNALTDYEHLADFIPNLVWSGRIPCPYP-----GRIWLEQRGFQRAMYWHIEARVVLDLQEVVNSAWDRELHFSMVDGDFKKFDGKWSVKS-GTRSSTAILSYEVNVIPRF---NFPAIFLERIIRSDLPVNLRALAYRAER Pt VWNSLTDYERLADFIPNLVCSGRIPCPHP-----GRVWLEQRGLQRALYWHIEARVVLDLQEFPHSANNRELHFSMVDGDFKKFEGKWSLRS-GTRHGTTTLSYEVNVMPRY---NFPAIFLERIIGSDLPVNLRALACRAER Bd VWRIITDYERLAEFVPNLVHSGRIPCPHE-----GRIWLEQRGLQQALYWHIEARVVLDLREVPDAVNGRELHFSMVDGDFKKFEGKWSVRS-GPRSASAILLYEVNVIPRF---NFPSIFLERIIRSDLPVNLRALAFRSEK Os VWRVITDYERLAEFIPNLVHSGRIPCPHQ-----GRVWLEQRGLQQALYWHIEARVVLDLKEVPDAVNGRELHFSMVDGDFKKFEGKWSIRS-GPRSSSAILLYEVNVIPRF---NFPAIFLERIIRSDLPVNLGALACRAEN Sb LWQVITDYERLADFIPNLVQSGTIPCPHE-----GRIWLEQRGLQQALYWHIEARVVLDLQEIHDSINGRELHFSMVDGDFKKFEGKWSIRS-GPRSSSAVLLYEVNVIPRF---NFPAIFLEKIIRSDLPVNLGALACRAEK Sm VWEVLTDYERLAEFIPNLIHSARIPCPYP-----GRIWLLQRGLHTAMYWHIEATVVLDLEEFPHLTDGRSLQFCMVDGDFKKYAGRWLLQA-GTRPGTTDLHYEVNVIPRL---LLPGVFVEGIIKSDLPVNLRAIAERAEK Pp VWEVLTDYGRLAEFIPNLTRSEQIPCPHP-----GRTWLLQEGKQSAMYWQIEARVVLDLEEFLDAKDGRELRFSMVDGDFKRYVGRWYLRP-DVRPGTIILHYEVNVTPRL---LFPAAFVEKIIKSDLPTNLRAIAARAED :*. :*** :**:*:*** * ****: ** ** *.* : *:**:*** *****.*..*:*.*******:: *:* ::. * * ***** ** :*. *:* ******* ** *:* ::* Cs VWAVLTDYDRLVEFVPNLEVCEKLPGGSA-----TRYRLRQQGCSQSLYLRLEASAVLDVQEVKGPLGRRELRFAMVESPNLKFSGQWTVEPDPTVRDGRSLGTTKLRYEIS---VAPKWSIPSTLVSKVVKSGLPANICAIA Cv VWRVLTNYERLADFVPNLESCERLPSPRT-----GRVWIRQRGCSQGVLWRLEAEAVIAVEEVRLPLGRREARFNMVDGDFKEMSGRWVVEPDP---SSAVGMATLLRFDIT---VQPKISLPSSVVSYVVRAGLPANIQAVS HCF145 SRPBCC2 ( AA) At VWKVLTSYESLPEIVPNLAISKILSRDNN----KVRI--LQEGCKGLLYMVLHARAVLDLHEIRE----QEIRFEQVEGDFDSLEGKWIFEQLGSHHTLLKYTVESKMRKDS---FLSEAIMEEVIYEDLPSNLCAIRDYIEK Gm VWNILTAYETLPKIVPNLAISKVVSRDNN----KVRI--LQEGCKGLLYMVLHARVVLDLCEYLE----QEISFEQVEGDFDSFRGKWIFEQLGNHHTLLKYSVESKMRKDT---FLSEAIMEEVIYEDLPSNLSAIRDYIEN Pt VWNVLTAYESLPEFVPNLAISKILSRENN----KVRI--LQEGCKGLLYMVLHARVVLDLCEHLE----QEISFEQVEGDFDSFQGKWILEQLGSHHTLLKYNVESKTHRDT---FLSEAIMEEVIYEDLPSNLCAIRDYIEK Bd VWNVLTAYE-LPEIIPNLAISRILLRDNN-----VRI--LQEGCKGLLYMVLHARVVM-LREKLE----REISFEQVEGDFFSFKGKWRLEQLGDQHTLLKYMVETKMHKDT---FLSESILEEVIYEDLPSNLCAIRDYVEK Os VWNILTAYEKLPEFVPNLAISRIIRRDNN----KVRI--LQEGCKGLLYMVLHARVVMDLREKLE----REISFEQVEGDFYSFKGKWRLEQLGDQHTLLKYMVETKMHKDT---FLSESILEEVIYEDLPSNLCAIRDYIEK Sb VWNVLTAYENLPEFVPNLAISRIVLRDNN----KVRI--MQEGCKGLLYMVLHARVVMDLREKFE----QEIRFEQVEGDFYSFKGKWRLEQLGDQHTLLKYMVETKMHRDT---FLSESILEEVIYEDLPSNLCAIRDYIEK Sm VWNVLTSYETLSEFVPNLSSSKIVSRHGN----HARV--LQEGCKCLLYMVLHARVVLELQELPP----NEITFQQVEGDFDVFSGKWTLESLGAEHTLLRYSVDMKMHNDF---LLPREIIEEIVYEDLPENLCAIRARVEL Pp VWAVLTAYESLQEFIPNLAICKVLTREKN----KVRL--LQEGCKCLLYMVLHARVILDLWERPQ----YEILFQQVEGDFDSFQGKWTLEPLGAQHTLLKYLVDTKMHKDS---LLAEALVEEVIYEDLPANLCAIRDRVEL **::**:** * :::***:.::: *. * :.*: :***** *********.:: * ** *:*.**** : *** :* **.****:* *: * :.* :*.. ::**::***** *:.*** :* Cs VWDVLTDYEALPEFVPNLAVCERLPVPAGMESRLTRL--RQVGFKDMVFMQLHAEAVLDLHERPH----REIQFRAVAGDFGVLQGKFMLSEPERKETHLKYAVEVKIPRSTPMMGLLEPILERMVYEDIPFNLAALKQR Cv VWDVLTDYNRLAEFIPNLAVSQRIALPSNAPANIIRI--RQVGYKRMLYMCLHAESVLDLIEKPQ----GEIQFRQVAGDFERFQGKWMLQGLPLSGNSSSTTSDAEPSASQTQLKYAVEIVIPRSTRMLGVLEPLLERT B RNA binding motif TMR1 (68 AA) At EVLKSEILKFISEHGQE-GFMPMRKQLRLHGRVDIEKAITRMGGFRRIALMMNLSLAYKHRKPKGYWDN Vascular plants: Gm KVLESELLKFIAEHGQE-GFMPMRKQLRLHGRVDIEKAITRMGGFRKIATILNLSLAYKHRKPKGYWDN At: Arabidopsis thaliana At5g08720 Pt DVLKSELLKFISEHGQE-GFMPMRKQLRLHGRVDIEKAITRMGGFRRIATLMNLSLAYKHRKPKGYWDN Gm: Glycine max XM_ Bd EVLKSELGSFISKYGQN-GFMPKRKHLRTHGRVDIEKAITRMGGFRKIASIMNLSLSYKNRKPRGYWDN Pt: Populus trichocarpa XM_ Os EVLKSELEKFIAKYGQD-GFMPKRKHLRLHGRVDIEKAITRMGGFRKIASIMNLSLSYKNRKPRGYWDN Bd: Brachypodium distachyon XM_ Sb EVLKSELENFIAEYGQY-GFMPKRKHLRSHGRVDIEKAITRMGGFRKIASIMNLSLSYKNRKPRGYWDN Os: Oryza sativa Os03g Sm KVLERELEGFVAKAGKE-RVMPVRAELRKNGRVDLEKAIRRFGGFRSIAERLNMSLAYKRRKPRGFWQN Sb: Sorghum bicolor XM_ Pp NILQQELLKFIAEKGTK-GVMPLRCELREAGRVDLEKAITRNGGFGPVASKLNLSLAYKERKPRGYWDN Sm: Selaginella moellendorffii XM_ :*: *: *::: *.** *.** ****:**** * *** :* :*:**:**.***:*:*:* Pp: Physcomitrella patens XM_ Cs EDFGLLAAELERCFGGT-GTMPTRAQLRAITRTDLEKAMVAHGGPAAVAKRMGWKLAYKAKAPRGYWDK Cv VGRKCRLPYLAFSPVLE-ALCPQKMTLYGAGRYDIARAVERWGGLYELAGELGYAVTGSRKPGFSEWQE Green Algae: HHHHHHHHHHHHHH HHHHHHH-HHHHHHHHHH---HHHHHHH Cs: Coccomyxa subellipsoidea XM_ RNA binding motif TMR2 (67-69 AA) Cv: Chlorella variabilis XM_ At ENLQEEIGRFQQSWGMDPSFMPSRKSFERAGRYDIARALEKWGGLHEVSRLLALNVRHPNRQLNSRKDN Gm ENLQEEISRFQRGWGIDPSFMPSRKSFERAGRYDIARALEKWGGLHEVSRLLSLKVRQRSRQDNLAKDK Pt ENLQEEISRFQRSWGMDLSFMPSRKSFERAGRYDIARALEKWGGLHEVSRLLALKVRHPNRQANSIKDR Bd ENLQEEI-RFQ-NWGIDPSYMPSRKSFERAGRYDIARALEKWGGIQEVSRLLSLEPRRPRRQADSDSEK Os ENLQEEIRRFQKNWGMDPAYMPSRKSFERAGRYDIARALEKWGGVHEVSRLLSLELRRPRRRANSDDES Sb ENLQEEISRFQKSWGMDPSYMPSRKSFERAGRYDIARALEKWGGVQEVSRLLSLKLRRPRRQGDLDDES Sm ENLKREIQLFQKKLRSDPSRMPSRRTLERAGRY-IARALEKWGGLHEVAKVLNLQTKRKRCLEEPEEGW Pp QNVHKEILLFQKEHGNDRTTMPTRQSLERAGRYDLARSLEKWGGLREVARVLGLQVKKRQKSRTAKTDV :*::.** ** * : **:*:::****** :**:******::**:::* *: :: Cs KNVEREIAEFCEQEGLPPRIMPLKMDFVRANRYDLAHVVERWGGLSELAELLEYQACTCRSVSPHVCKE CV HISELAASTGLSGREGLFELASKTYAARRSMQGSVDGGEDMALADILTADAVGARAASANSKASAENGA HHHHHHHHHHHHHH HHHHHHH-HHHHHHHHHH---HHHHHHH C 5
6 Supplemental Figure 4. Conservation of the Tandem Repeated SRPBCC and TMR motifs in HCF145. (A) Alignment of tandem repeated HCF145 motifs SRPBCC1 and SRPBCC2. The number of amino acids (aa) is indicated. Acidic and basic amino acids are labeled in red and blue, respectively. Highly conserved amino acids in both motifs are boxed in yellow. (B) Alignment of tandem repeated RNA binding TMR motifs 1 and 2. The number of amino acids (aa) is indicated. Highly conserved amino acids in both TMR motifs are boxed in yellow. The predicted helices (Jpred [ and NetSurfP [ are shown below the alignment. For schematic presentation see Figure 3. (C) Abbreviations of organisms and accession numbers used in the alignments. 6
7 A Np: Csp: Cst: Ds: Gs: Pt: Vv: At: Os: Bd: Sb: Si: Sm: Pp: Cr: Cs: Pm: Ss: AtM1: AtM2: Ct: Pa: EKIWKVLTDYEALPDFLPNLAKSRLIEHPNGG-IRLEQVGSQ-RLLNFNFSARVVLDLEECFP REINFR-MVEGDFKGFSGSWCLEPYSLG EYIGTNLCYTIQV--WPKLTMPVGIIENRLSKDLRLNL EKTWQVLTDYEALADFIPNLIKSRLLEHPDGG-IRLEQIGSQ-RLLNFNFCARVVLDLEEYFL KEINFR-MIEGDFKGFSGSWCLKPYSFG DLVGTDLCYTIQV--WPKLTMPLKIIEPRLTNDMHVNL EKIWQILTDYESLADFIPNLAQSRLLAHPQGG-IRLEQIGSQ-RLLNFKFCARVVLDLEELFP KEINFQ-MVEGDFKGFSGKWCLEPYSLG AAQGTNLCYTIQV--WPKLTMPISILENRLSNDLRLNL EAVWQVLTDYESLPEFIPSLEKSQRLEHPEGEKVRLEQVGKQ-RLFKVNFSARVVLDLTEMPP SRIDFE-MVEGDFKAFSGYWSLEEADQ KTELIYSIFV--WPPRTMPVSLIERRLSLDLSLNL KAVWDLLTDYEHLAEFIPNLAVSRLRYHPQGG-IRLEQEGVQ-SVLGFRFRASVILDMYEKFS EDRAEIDFVLADSQDFDVFEGSWLMYPMKR NWTHLIYQVTV--QPKRFVPVQAVEWRIREDVPSNL DTVWKILTDYEKLADFIPGLAVSKLIDKKDKF-ARLYQIGQQNLAFGLKFNAKAILDCYERDLQTLAS----GEKRDIEFK-MTEGDFQFFEGMWSIEQLAK----PKTEDSVGQEYETTLSYLVDV--KPKMWLPVNLIEGRICKEIKSNL HTVWSILTDYEGLADFIPGLAVSQLVEKGEKF-ARLFQIGQQDLAFGLKFNAKGIVDCYEKDLESLPF----GEKRDIEFK-MIEGDFQIFEGKWSIEQRNTNT-WEGKDSSVGQEFYTTLTYVVDV--EPKRWLPVYLVEGRLSREIKMNL DSVWSVLTDYEKLSDFIPGLVVSELVEKEGNR-VRLFQMGQQNLALGLKFNAKAVLDCYEKELEVLPH----GRRREIDFK-MVEGDFQLFEGKWSIEQLDKGI-HGEALDLQFKDFRTTLAYTVDV--KPKMWLPVRLVEGRLCKEIRTNL DAVWATLTDYEGLAGFIPGLSECRLLDQSDCF-ARLYQVGEQDLALGFKFNARGTIDCYEGELQLLP---AGARRREIAFN-MIDGDFKVFEGNWSVQEEVD-----GGEISADQEFQTILSYVVEL--EPKLWVPVRLLEGRICNEIKTNL EAVWATLTDYEGLAGFIPGLSECRLLHQDAAF-ARLYQVGEQDLALGFKFNAKGTIDCYEGEMEVLP---AGARRREIAFN-MVEGDFKVFEGKWSVEEVEDSL-DEGGENPTGQEFQTTLSYVVEL--EPKLWVPVRLLEGRICKEIKTNL EAVWATLTDYEGLADFIPGLSECRLLDQHDGF-ARIYQVGEQDLALGFKFNAKGTIDCYEGDMEVLPD--AGARRREIAFN-MIDGDFKLFQGKWSVEEVDGSI-VEGGGNSEEQEFQTTLSYLLEL--EPKLWVPVRLLEGRICSEIKNNL EAVWATLTDYEGLADFIPGLSECRLLDQAQGF-ARLYQVGEQDLALGFKFNAKGTIDCYEGDLESLPDAQGNARRREIAFN-MIDGDFKVFQGKWSVQESVEQEQGGGDSDEGQESQTTTLSYLVEL--EPKLWVPVRLLEGRICSEIKNNL ETVWGVLTDYEGLADFIPGLASSKVLERRENG-AQLLQIGEQELALGVKFRAKGVIEVTELPLELLDNG----CRRDIGFD-MVEGDFNLFRGIWRIEQILHG VEDATTQTSLTYILEV--QPKIWIPVALLEGRLQKEVSNNL EAVWGVLTDYDHLADHIPGLAESSVLQRRSNG-ARLKQIGQKNFALGVKFKAKAVVEVTEEAAQDLDDG----TLRDLHFE-TVEGDFQVFKGTWRMLEKSLE SNDAKVETYLSYILEV--QPKRWMPVALIEGVLGQEITCNL SAVWLALSDYDNLGKFIPSLVENRCLERGGRT-AVLYQVGAQDVAMGVKFSA-ALASVEALFP-YPLTSAPGVSSSDITFE-LVEGDFQAFRGVWRMQQT GEATTLLSYALFV--KPQAWLPVALIQGRIENEVVRNL EVIWGALTDYDSLGTFIPGLAENRCLERRAQG-AQLLQIGEQEIAFGAKFRARVVLDIEEHWSGVPG 35 aa HDIAFC-ACEGDFQVFRGVWRIQEGSR GEGSSRLSYALFV--RPQIWLPVRLVQGRIESEIKNNL DSLWSVLTDYDRLNLYIPNLLSSKKIFQKGNN-VHLKQVGAQ-DFLGMKFSAEVTIDLFEN KELGLLKFN-LIKGDFRKFEGSWKIQNIKNTS TNSLIYDLTV--QGCQWMPIGMIEKRLKKDLSENL DELWEVLTDYENLSKFIPNLSSSQLVHREGHT-VRLQQVGSQ-QLLGLRFSAQVQLELTEF RSEGLLSFK-MVKGDFRRFEGAWRVNELADG CSLVYELTV--QGCIGMPIALIEERLRDDLSSNL QSVWNVLTDYERLADFIPNLVWSGRIPCPHPGRIWLEQRGLQ-RALYWHIEARVVLDLHECLDSPNG RELHFS-MVDGDFKKFEGKWSVKSGIR SVGTVLSYEVNV--IPRFNFPAIFLERIIRSDLPVNL CEVWKVLTSYESLPEIVPNLAIS-KILSRDNNKVRILQEGCK-GLLYMVLHARAVLDLHEIRE QEIRFE-QVEGDFDSLEGKWIFEQ-LG SHHTLLKYTVESKMRKDSFLSEAIMEEVIYEDLPSNL KHVWAAITDYNNHKSFVPKLIDSGLISDNGRE-QVMFERGKTGIFLFRKTVYIKLSLQGEY PKRLDFH-QIEGDFKVYEGDWLIERASDG KGSILTFRAKIKPDFFAPAMFVRKVQQNDLPMVL ETIWNLLTDYNNLSTIIPKVIDSRLIEDNGSH-KIIDQTGKSGILFIEKSVRIVLKVTEKF PNALLFE-MVEGDFSTYTGSWSFRPGSSR EQTFVSWQTDFKPTFFAPPFLVSFLQHQDLPVVM * *:.*: : :*.: : * * : : : :. : *.. ** : * *. : : :.. :: : :: ** B AtM1: Arabidopsis thaliana HCF145 SRPBCC1 AtM2: Arabidopsis thaliana HCF145 SRPBCC2 Np: Nostoc punctiforme PCC73102 Csp: Calothrix sp. PCC7507 Cst: Cylindrospermum stagnale PCC7417 Ds: Dactylococcopsis salina PCC8305 Gs: Galdieria sulphuraria Pt: Populus trichcarpa Vv: Vitis vinifera At: Arabidopsis thaliana Os: Oryza sativa Bd: Brachypodium distachyon Sb: Sorghum bicolor Si: Setaria italica Sm: Selaginella moellendorffii Pp: Physcomitrella patens Cr: Chlamydomonas reinhardtii Cs: Coccomyxa subellipsoidea Pm: Prochlorococcus marinus sp. AS9601 Ss: Synechococcus sp. CC9902 Ct: Chlorobium tepidum Pa: Prosthecochloris aestuarii C At5g08720 At5g08720 CP CP CP CP XM_ XM_ XM_ At4g01650 AK XM_ XM_ XM_ XM_ XM_ XM_ XM_ CP CP AE CP
8 Supplemental Figure 5. Conservation of the SRPBCC Motif in HCF145-L Proteins of Photosynthetic Organisms. (A) Multiple alignment of HCF145-L proteins in photosynthetic organisms. HCF145 motifs SRPBCC1 (AtM1) and SRPBCC2 (AtM2) are included. Acidic and basic amino acids are labeled in red and blue, respectively. Highly conserved amino acids are boxed in yellow. For schematic presentation see Figure 3. (B) Abbreviations of organisms and accession numbers used in the alignment. (C) Unrooted phylogram of all sequence motifs used in the alignment analyzed by the maximum likelihood method. Bootstrap values (based on 1000 iterations) are shown for corresponding nodes. The scale bar is an idicator for the evolutionary distance in substitutions per site. 8
9 9
10 Vascular Plants Arabidopsis thaliana HCF145 At5g FMPMRKQLRLHGRVDIEKAIT--RMGGFRRIALMMNL-SLAYKHRKPKGYWD 2. FMPSRKSFERAGRYDIARALE--KWGGLHEVSRLLAL-NVRHPNRQLNSRKD Green algae Chlorella variabilis XM_ Chlorellales 1. TMPTSAQLEAAGRRDLVAAVRA--AGGFLEVAQALGLR----SQRKPAGYWE 2. VMPSRSALQAAGRYDLHHAVML--HGGYTVASQSLDR RPAWP 3. CLPTASQLLEAGRGDLYQASRW--GGGAIVRRGGFCA AGQALGWE 4. RMPTHLQLASAGRHDLKYALQL--HGSASIAAM-LGLQ------GNTQGAHN Bathycoccus prasinos CCO17813 Mamiellales 1. GMPSKRSLEKENRKDLIKRVEK--LFGYDWLTMAVLLDF--EPFRKPFYYWD 2. VMPTRRDLIDARRWDLHHAVVL--HGGYGAVAKTLKWPR--ARWAEDRHLLN 3. RLPSALELRNVGRDDLARHMVE--HGGPVTVAKRMRMKP GKGAWI 4. YMPTDEELINAGRHDLRYRVKE--IGSATVAKYAKLQNRTEKMSLAEARAFL Coccomyxa subellipsoidea C-169 XP_ Coccomyxaceae 1. RMPSCTELREAGAFTLYSAISK--HGGVGAFARQLGLDP----KRRDSGYWE 2. GMPTIQDLQRSGHNSLIKAINH--WGGRSAVARRLGLACSPTRRLMTLGDLS 3. VMPSRTQLLEAGRPDLLQAVKR--MGGFKRVAAALELAF----LPARRGRSA Ostreococcus lucimarinus XM_ Mamiellophyceae 1. CMPTREQLR-GGRHWDAIQQIE-SLGGFVKVAQLLDWSG---AKTRPRGYWT 2. RMPSQKSLRDAGRADIVNALKR--FGGAEKVAASMGLEFGSGNKRSSASARG Red algae Cyanidioschyzon merolae XM_ Cyanidiales TMPTAGQLAAHHRSDLIRAIRK--HGGFPKVAEQLGLK----AHRRPNGYWN Cyanidioschyzon merolae XM_ Cyanidiales VMPPERILAKAGRFDLIISIEY--HGGSRAVAEICELRDSASWEYVLEMRDL EMPSIAELQRQGREDLARLIRR--HGGPLVFAARFGLYVPPSRRRREADLKW Cyanidioschyzon merolae XM_ Cyanidiales 1. YMPTSNELREAKRSDLVRAIIV--HGGYAKVAERCGL--QPHR--RSFGYWR 2. RMPTYNELVAAKKRILAYAIAE--NGGFLEVARRMNLQLSSDE-----TPWR 3. RFPRQQDLVRLGRYDLDWAIHRW-HGGYTRLAAELGYLRSRLPC-KPRNFWS 4. RMPNRKELEALDRHDLIYAIRK--FGGFLTVATKLGLSRDALTHTRPRGYWS 5. IMPRLEQLRMYNREDLINAIHR--HGGAANVARRLHLFWY-----GPKTFWR 6. KMPTQQELISAGRVDVAYGVHL--HGGVYEVARRLRLQVLDPP--RAPFYWN 7. VMPTSMTIVRSGRRDLAAAIRR--HGGWDAFARRLNLRPAAPK--RPKGYWN 8. FVRNYAADFVRRPGDDEDDPDAAGKIAYSDVSEILAGNVGNKRCRTVVGAIR 9. VMPTAEELRLDGRADLVFACERI-HGGLATVARGLGWPLLAER--LPPESLK 10. EMPTEADLLRTGGIDIHEAIVC--HGGYVEVARSLNLRHPEDP---EWTDWS Galdieria sulphuraria XM_ Cyanidiales 1. SVHLLQVKKSGGECPLPRSSTKCYYGGGTPSFELLVP-20aa-VRDVRGYWK 2. RMPSANELRKSGRYALALAISSH-HGGFHAVAREIGLQPNHHS----SGYWD 3. YMPTYHQLLKARRLDLAKAIHK--YGGFPAVAEKLGR---IPN--KRRKYWH 4. QLPTMTLLSTCKRWDLMGAIRL--HGGLYEVSRKTQIPLSKSTR-QPRGYWS 5. IVPTLSNLKRNQRQDLVEAIRK--HGGVQTVAAKLYMLRQSKR--KAKGYWN 6. VMPLGHELRRHQRRDLCYAIQL--HGGFSVVAGKLHL---NWI--GPISFWR 7. RMPTLNDLVIRGRVDLAFGIRL--HHGFPAVAKAFGLEWTIPS--RPRMYWN 8. YMPSNETLYQLGRGDLADAIRD--TKGWVYYAKRLGLVPHYRCI-SSHKLWK 9. AVASVEELYRDGRGDIAFAIMKY-HQGATQLAHRLKWKAPHMRP-LPPAYYR 10. LMPSKKELFQTGYRDLVFVIYR--HGGFQKVACRMGWTIHEDNP-HWLTQWL Cyanobacteria Microcoleus sp. PCC 7113 AFZ22266 Oscillatoriales 1. VMPKAAQLRQLGRYDLA-MAISKYHGGYRSVASRLGLTYT----GQRFGYWH 2. VMPSRQQLEQAGEKPLAAAIG-LHGGVL-AVARRLGFKLP--YGRKPRGYWK 3. VMPTREQLVQIQRAELISAIA-TNGGWP-SVARRFGL------ANPNKGYTS 10
11 Supplemental Figure 7. Multiple Alignment of Transcript Binding Motif Repeats (TMR) Present in Representative TMR Proteins of Photosynthetic Organisms in Eukaryotes and Cyanobacteria. The consensus sequence MP-x(4)-L-x(3)-GR-x-DL-x(2)-AI-x(2-4)-HGG-x(3)- VA-x(2)-LGL-x(5-27)-GYW is colored in yellow. Acidic and basic amino acids are labeled in red and blue, respectively. For schematic presentation see Figure 3. 11
12 12
13 Supplemental Table 1. Oligonucleotides Used for PCR, RT-PCR, Primer Extension, Probe Generation, Genotyping, and other Applications. Primer name Primer sequence ATG-f-P 720-3UTR-r-P Cacc-145-for 145-rev. ex7-for ex8-for ex10-rev2 ex10-rev1 LBb1 PsaA80-mer At4-Kpn1-f At4-Kpn1-r Cacc-145-for 145-rev Fw-BclI-hcf145 Rev-SalI-hcf145-strep Rev-SalI-hcf145- strepa2 Fw-BclI-hcf145-B1 Rev-SalI-hcf145-strep psaa ATG rev 70-mer psaa 1 70-mer psaa 2 70-mer psaa 3 psba 80-mer T7_5UTRpsaA 3 T7_5UTR-psaA 2 psaa5utrrev HCF145-like 2-f HCF145-like 2-r LB4 B3-T7-petB-for petb-ex-rev atgtcagtgagcaagtttccacatctc tatctttgcgagttacaagactacac caccatgtcagtgagcaagtttccacatctc atattgaacccaattgatatcaagatc ctgaagcaatcatggaagagg agctttgatgatgaatctttcacttgc ggatgtctcacgttcaatgc aagacgagatacttcgtgtaatcctc gcgtggaccgcttgctgcaact ccacatctccattcaggatttcttggcccactattggccaaaccacctgagcactaggtccaatgtgagtagg atcactc ctgctttggtaccgctttccagatctag atcatggtacccaaccaattcgctaacaagacc caccatgtcagtgagcaagtttccacatctc atattgaacccaattgatatcaagatc atatattgatcacatagcggcgccggtag ataatagtcgacttatttttcgaactgcgggtggctccaagcgctatattgaacccaattgatatcaag ataatagtcgacttatttttcgaactgcgggtggctccaagcgctaatgtctctttgtagaccaggaat atatattgatcaagaggagagaaatcttcagag ataatagtcgacttatttttcgaactgcgggtggctccaagcgctatattgaacccaattgatatcaag gttccggcgaacgaataatcat tctattttctttagttattcactagagcaattatgatctggaagtcgatctggggcaagtgttcggatct aagtaattagtgaaccgatgatagatacttagaatttttttttctttctcttctatcttccatctattca gcgaacgaataatcattgagtcctcctctttccggacaacacatacaaagaaacccgccaacagtcactc gacggttttcagtgctagttatccagttacagaagcgaccccataggctttcgctttcgcgtctctctaaaattg cagtc taatacgactcactatagggagaagatccgaacacttgccc taatacgactcactatagggctaagtatctatcatcggttcac gtgaaccgatgatagatacttag ctcgacactgctagcttcttc tcgtaacaatctagaacagctttg cgtgtgccaggtgcccacggaatagt cgtaatacgactcactatagggactgaagctaactttgg taccggaatagcgtcaggtacac 13
14 Supplemental Table 1. continued Primer name Primer sequence 5 3 psaa 5' 1 pppsaa_f pppsaa_r pppsba_f pppsba_r pprps14_f pprps14_r ppycf3_f ppycf3_r npt5 npt3 pp5 f pp5 r pp3 f pp3 r ppf1 ppr1 ppf2 ppr2 ppf3 ppr3 EF1α-f EF1α-r Fw T7 psaa3_1 Fw T7 psaa3_2 Rev psaa3_1 Rev psaa3_2 taccggaatagcgtcaggtacac tggcgagcatctggaataacta tgtccagcaaccagaaaaagaa ttcattgcagctcctcctgtag aggatgttgtgttcagcttgga aaaaatatcaagattttcgtcactc tgcatgagccatttctcgta tgttgcagacattttactccgaat ccaatgctattgcttgtttcca agtgcattctttggacctccaatcggatcctgtcaaacactga gacaggaggcccgatctagtaa gctcgagtttttcagcaagatgtggccaacacaaatgaacagt tggaggtccaaagaatgcacta ttactagtcgggcctcctgtccacgtgtttcttgaaccagagg aggagatcttctagaaagatgtcgacctcatatgaatggccacgatgt atcagcagtgattctgccctaag gcggctgagtggctccttca aaattatcgcgcgcggtgtc tgatcatggtgagaagtgttgct ggtttgggaggttttgactgac cgtggagagacaacatcctttg agcgtggtatcacaattgac gatcgctcgatcatgttatc taatacgactcactatagggagacataattgctctagtgaataactaa taatacgactcactatagggagagaatagatggaagatagaagagaa ttagttattcactagagcaattatg ttctcttctatcttccatctattc 14
tgg agg Supplementary Figure S1.
 ttaggatattcggtgaggtgatatgtctctgtttggaaatgtctccgccattaactcaag tggaaagtgtatagtaatgaatctttcaagcacacagatcacttcaaaagactgtttcaa catcacctcaggacaaaaagatgtactctcatttggatgctgtgatgccatgggtcacag attgcaattcccaagtgcccgttcttttacaccaaaatcaaagaagaatatctccccttt
ttaggatattcggtgaggtgatatgtctctgtttggaaatgtctccgccattaactcaag tggaaagtgtatagtaatgaatctttcaagcacacagatcacttcaaaagactgtttcaa catcacctcaggacaaaaagatgtactctcatttggatgctgtgatgccatgggtcacag attgcaattcccaagtgcccgttcttttacaccaaaatcaaagaagaatatctccccttt
Supporting information
 Supporting information Figure S1. Carotenoid biosynthesis pathway in papaya fruit which is adopted from Blas et al. (2010) (reference 4) and Nisar et al. (2015) (reference 5). Carotenoids are synthesized
Supporting information Figure S1. Carotenoid biosynthesis pathway in papaya fruit which is adopted from Blas et al. (2010) (reference 4) and Nisar et al. (2015) (reference 5). Carotenoids are synthesized
Experimental Identification and Computational Characterization of a Novel. Extracellular Metalloproteinase Produced by Clostridium sordellii
 Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 207 Supplementary Information Experimental Identification and Computational Characterization of
Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 207 Supplementary Information Experimental Identification and Computational Characterization of
Strain or plasmid Description a Reference or source b. 168 trpc2 Laboratory. 1A780 trpc2 sigb::spc BGSC
 TABLE S1 Bacterial strains and plasmids Strain or plasmid Description a Reference or source b B. subtilis 168 trpc2 Laboratory stock 1A780 trpc2 sigb::spc BGSC BM1010 trpc2 htpx::pgs1582; Em r BM1302 trpc2
TABLE S1 Bacterial strains and plasmids Strain or plasmid Description a Reference or source b B. subtilis 168 trpc2 Laboratory stock 1A780 trpc2 sigb::spc BGSC BM1010 trpc2 htpx::pgs1582; Em r BM1302 trpc2
FETAL FIBROBLASTS, PASSAGE 10
 Double-stranded methylation patterns of a 104-bp L1 promoter in DNAs from fetal fibroblast passages 10, 14, 17, and 22 using barcoded hairpinbisulfite PCR. Fifteen L1 sequences were analyzed for passages
Double-stranded methylation patterns of a 104-bp L1 promoter in DNAs from fetal fibroblast passages 10, 14, 17, and 22 using barcoded hairpinbisulfite PCR. Fifteen L1 sequences were analyzed for passages
MALE ADULT FIBROBLAST LINE (82-6hTERT)
 Double-stranded methylation patterns of a 104-bp L1 promoter in DNAs from male and female fibroblasts, male leukocytes and female lymphoblastoid cells using hairpin-bisulfite PCR. Fifteen L1 sequences
Double-stranded methylation patterns of a 104-bp L1 promoter in DNAs from male and female fibroblasts, male leukocytes and female lymphoblastoid cells using hairpin-bisulfite PCR. Fifteen L1 sequences
enzymatic families regulated in response to a short-term cytokinin treatment in root apices
 A B C D Supplemental tlfigure 1. 1Mtbli Metabolic processes, hormonal pathways and dlarge enzymatic families regulated in response to a short-term cytokinin treatment in root apices The 297 transcripts
A B C D Supplemental tlfigure 1. 1Mtbli Metabolic processes, hormonal pathways and dlarge enzymatic families regulated in response to a short-term cytokinin treatment in root apices The 297 transcripts
Tomato SlDREB gene restricts leaf expansion and internode elongation by down-regulating key
 Title: Tomato SlDREB gene restricts leaf expansion and internode elongation by down-regulating key genes for gibberellin biosynthesis Authors: Jinhua Li 1, Wei Sima 1, Bo Ouyang 1, Taotao Wang 1, Khurram
Title: Tomato SlDREB gene restricts leaf expansion and internode elongation by down-regulating key genes for gibberellin biosynthesis Authors: Jinhua Li 1, Wei Sima 1, Bo Ouyang 1, Taotao Wang 1, Khurram
Methods S1. Sequences relevant to the constructed strains, Related to Figures 1-6.
 Methods S1. Sequences relevant to the constructed strains, Related to Figures 1-6. A. Promoter Sequences Gal4 binding sites are highlighted in the color referenced in Figure 1A when possible. Site 1: red,
Methods S1. Sequences relevant to the constructed strains, Related to Figures 1-6. A. Promoter Sequences Gal4 binding sites are highlighted in the color referenced in Figure 1A when possible. Site 1: red,
Eukaryotic Comparative Genomics
 Eukaryotic Comparative Genomics Detecting Conserved Sequences Charles Darwin Motoo Kimura Evolution of Neutral DNA A A T C TA AT T G CT G T GA T T C A GA G T A G CA G T GA AT A GT C T T T GA T GT T G T
Eukaryotic Comparative Genomics Detecting Conserved Sequences Charles Darwin Motoo Kimura Evolution of Neutral DNA A A T C TA AT T G CT G T GA T T C A GA G T A G CA G T GA AT A GT C T T T GA T GT T G T
Underpinning Phage Arbitrium Communication Systems
 Molecular Cell, Volume 74 Supplemental Information Deciphering the Molecular Mechanism Underpinning Phage Arbitrium Communication Systems Francisca Gallego del Sol, José R. Penadés, and Alberto Marina
Molecular Cell, Volume 74 Supplemental Information Deciphering the Molecular Mechanism Underpinning Phage Arbitrium Communication Systems Francisca Gallego del Sol, José R. Penadés, and Alberto Marina
Chapter 7. Motif finding (week 11) Chapter 8. Sequence binning (week 11)
 Course organization Introduction ( Week 1) Part I: Algorithms for Sequence Analysis (Week 1-11) Chapter 1-3, Models and theories» Probability theory and Statistics (Week 2)» Algorithm complexity analysis
Course organization Introduction ( Week 1) Part I: Algorithms for Sequence Analysis (Week 1-11) Chapter 1-3, Models and theories» Probability theory and Statistics (Week 2)» Algorithm complexity analysis
Supplementary information: Biocatalysis on the surface of Escherichia coli: melanin pigmentation of the cell. exterior
 Supplementary information: Biocatalysis on the surface of Escherichia coli: melanin pigmentation of the cell exterior Martin Gustavsson, David Hörnström, Susanna Lundh, Jaroslav Belotserkovsky, Gen Larsson
Supplementary information: Biocatalysis on the surface of Escherichia coli: melanin pigmentation of the cell exterior Martin Gustavsson, David Hörnström, Susanna Lundh, Jaroslav Belotserkovsky, Gen Larsson
Changes in the drawing are allowed only by the permission of the authorities who have granted the certificate Muutokset sallittu vain sertifikaatin my
 Changes in the drawing are allowed only by the permission of the authorities who have granted the certificate Muutokset sallittu vain sertifikaatin myöntäjän luvalla The drawing is a valid document only
Changes in the drawing are allowed only by the permission of the authorities who have granted the certificate Muutokset sallittu vain sertifikaatin myöntäjän luvalla The drawing is a valid document only
Plasmid Name: pmm290. Aliases: none known. Length: bp. Constructed by: Mike Moser/Cristina Swanson. Last updated: 17 August 2009
 Plasmid Name: pmm290 Aliases: none known Length: 11707 bp Constructed by: Mike Moser/Cristina Swanson Last updated: 17 August 2009 Description and application: This is a mammalian expression vector for
Plasmid Name: pmm290 Aliases: none known Length: 11707 bp Constructed by: Mike Moser/Cristina Swanson Last updated: 17 August 2009 Description and application: This is a mammalian expression vector for
Changes in the drawing are allowed only by the permission of the authorities who have granted the certificate Muutokset sallittu vain sertifikaatin my
 Changes in the drawing are allowed only by the permission of the authorities who have granted the certificate Muutokset sallittu vain sertifikaatin myöntäjän luvalla The drawing is a valid document only
Changes in the drawing are allowed only by the permission of the authorities who have granted the certificate Muutokset sallittu vain sertifikaatin myöntäjän luvalla The drawing is a valid document only
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
Functional Genomics & Proteomics
 Functional Genomics & Proteomics Genome Sequences TCACAATTTAGACATCTAGTCTTCCACTTAAGCATATTTAGATTGTTTCCAGTTTTCAGCTTTTATGACTAAATCTTCTAAAATTGTTTTTCCCTAAATGTATATTTTAATTTGTCTCAGGAGTAGAATTTCTGAGTCATAAAGCGGT CATATGTATAAATTTTAGGTGCCTCATAGCTCTTCAAATAGTCATCCCATTTTATACATCCAGGCAATATATGAGAGTTCTTGGTGCTCCACATCTTAGCTAGGATTTGATGTCAACCAGTCTCTTTAATTTAGATATTCTAGTACAT
Functional Genomics & Proteomics Genome Sequences TCACAATTTAGACATCTAGTCTTCCACTTAAGCATATTTAGATTGTTTCCAGTTTTCAGCTTTTATGACTAAATCTTCTAAAATTGTTTTTCCCTAAATGTATATTTTAATTTGTCTCAGGAGTAGAATTTCTGAGTCATAAAGCGGT CATATGTATAAATTTTAGGTGCCTCATAGCTCTTCAAATAGTCATCCCATTTTATACATCCAGGCAATATATGAGAGTTCTTGGTGCTCCACATCTTAGCTAGGATTTGATGTCAACCAGTCTCTTTAATTTAGATATTCTAGTACAT
I. Principles of Pointer Year Analysis
 I. Principles of Pointer Year Analysis Fig 1. Maximum (red) and minimum (blue) pointer years. 1 Fig 2. Principle of pointer year calculation. Fig 3. Skeleton plot graph created by Kinsys/Kigraph programme.
I. Principles of Pointer Year Analysis Fig 1. Maximum (red) and minimum (blue) pointer years. 1 Fig 2. Principle of pointer year calculation. Fig 3. Skeleton plot graph created by Kinsys/Kigraph programme.
Supporting Information for
 Supporting Information for Analysis of Sogatella furcifera proteome that interact with P10 protein of Southern rice black-streaked dwarf virus Win Than*, Faliang Qin*, Wenwen Liu, Xifeng Wang ** State
Supporting Information for Analysis of Sogatella furcifera proteome that interact with P10 protein of Southern rice black-streaked dwarf virus Win Than*, Faliang Qin*, Wenwen Liu, Xifeng Wang ** State
11/17/11. Gene Regulation. Gene Regulation. Gene Regulation. Finding Regulatory Motifs in DNA Sequences. Regulatory Proteins
 Gene Regulation Finding Regulatory Motifs in DNA Sequences An experiment shows that when X is knocked out, 20 other s are not expressed How can one have such drastic effects? Regulatory Proteins Gene X
Gene Regulation Finding Regulatory Motifs in DNA Sequences An experiment shows that when X is knocked out, 20 other s are not expressed How can one have such drastic effects? Regulatory Proteins Gene X
rapid evolution in copy number, location and sequence, with diverse turnover mechanisms.
 Repetitive DNA and its evolution: genomic strategies to understand the processes and exploit biodiversity rapid evolution in copy number, location and sequence, with diverse turnover mechanisms. often
Repetitive DNA and its evolution: genomic strategies to understand the processes and exploit biodiversity rapid evolution in copy number, location and sequence, with diverse turnover mechanisms. often
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
Computational personal genomics: selection, regulation, epigenomics, disease
 Computational personal genomics: selection, regulation, epigenomics, disease Manolis Kellis Broad Institute of MIT and Harvard MIT Computer Science & Artificial Intelligence Laboratory TATTGAATTTTCAAAAATTCTTACTTTTTTTTTGGATGGACGCAAAGAAGTTTAATAATCATATTACATGGCATTACCACCATATA
Computational personal genomics: selection, regulation, epigenomics, disease Manolis Kellis Broad Institute of MIT and Harvard MIT Computer Science & Artificial Intelligence Laboratory TATTGAATTTTCAAAAATTCTTACTTTTTTTTTGGATGGACGCAAAGAAGTTTAATAATCATATTACATGGCATTACCACCATATA
RUOSTUMA TTOMA RUOSTUMATTOMAT 86-103 RUOSTUMATTOMAT
 RUOTUMATTOMAT RUOTUMATTOMAT... 86-103 RUOTUMATTOMAT Levyt... 88-92 Kelat... 92 Tangot... 93-97 Putket... 98-100 Tekniset tiedot... 101-102 Tuotantopalvelut... 102 Olee jatkuvasti kehittäneet tuotevalikoimaae
RUOTUMATTOMAT RUOTUMATTOMAT... 86-103 RUOTUMATTOMAT Levyt... 88-92 Kelat... 92 Tangot... 93-97 Putket... 98-100 Tekniset tiedot... 101-102 Tuotantopalvelut... 102 Olee jatkuvasti kehittäneet tuotevalikoimaae
6.095/ Computational Biology: Genomes, Networks, Evolution. Sequence Alignment and Dynamic Programming
 6.095/6.895 - Computational Biology: Genomes, Networks, Evolution Sequence lignment and Dynamic Programming Tue Sept 13, 2005 Challenges in Computational Biology 4 Genome ssembly 5 Regulatory motif discovery
6.095/6.895 - Computational Biology: Genomes, Networks, Evolution Sequence lignment and Dynamic Programming Tue Sept 13, 2005 Challenges in Computational Biology 4 Genome ssembly 5 Regulatory motif discovery
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
Supporting Information for Electrophoresis DOI /elps Sunil Archak, Vemireddy Lakshminarayanareddy and Javaregowda Nagaraju
 ELECTROPHORESIS Supporting Information for Electrophoresis DOI 10.1002/elps.200600646 Sunil Archak, Vemireddy Lakshminarayanareddy and Javaregowda Nagaraju High-throughput multiplex microsatellite marker
ELECTROPHORESIS Supporting Information for Electrophoresis DOI 10.1002/elps.200600646 Sunil Archak, Vemireddy Lakshminarayanareddy and Javaregowda Nagaraju High-throughput multiplex microsatellite marker
Predicting evolutionarily conserved regions (ECRs) in the Xenopus tropicalis genome using a MultiPipMaker-based bioinformatic strategy
 Predicting evolutionarily conserved regions (ECRs) in the Xenopus tropicalis genome using a MultiPipMaker-based bioinformatic strategy OVERVIEW- Method updated and written by Sarah Louie (6/19/08) Initially
Predicting evolutionarily conserved regions (ECRs) in the Xenopus tropicalis genome using a MultiPipMaker-based bioinformatic strategy OVERVIEW- Method updated and written by Sarah Louie (6/19/08) Initially
Määräys STUK SY/1/ (34)
 Määräys SY/1/2018 4 (34) LIITE 1 Taulukko 1. Vapaarajat ja vapauttamisrajat, joita voidaan soveltaa kiinteiden materiaalien vapauttamiseen määrästä riippumatta. Osa1. Keinotekoiset radionuklidit Radionuklidi
Määräys SY/1/2018 4 (34) LIITE 1 Taulukko 1. Vapaarajat ja vapauttamisrajat, joita voidaan soveltaa kiinteiden materiaalien vapauttamiseen määrästä riippumatta. Osa1. Keinotekoiset radionuklidit Radionuklidi
Fungi infecting cultivated moss can also cause diseases in crop plants
 Fungi infecting cultivated moss can also cause diseases in crop plants SEMINAR ON PEST RISK ASSESSMENT 2-3 October 2012, Helsinki, Finland 9.10.2012 1 Fungi infecting cultivated moss can also cause diseases
Fungi infecting cultivated moss can also cause diseases in crop plants SEMINAR ON PEST RISK ASSESSMENT 2-3 October 2012, Helsinki, Finland 9.10.2012 1 Fungi infecting cultivated moss can also cause diseases
CSC:n käyttäjätunnukset - myös opiskelijoille
 CSC:n käyttäjätunnukset - myös opiskelijoille http://www.csc.fi/asiakkaaksi/korkeakoulut/kayttol upahakemukset/index_html Ohjaajan nimeksi Petri Törönen Perusteluiksi opiskelu ja luentokurssin nimi (Geneettisen
CSC:n käyttäjätunnukset - myös opiskelijoille http://www.csc.fi/asiakkaaksi/korkeakoulut/kayttol upahakemukset/index_html Ohjaajan nimeksi Petri Törönen Perusteluiksi opiskelu ja luentokurssin nimi (Geneettisen
Contents. Kuinka monta jakson sanaa opit? Väritä tähdet. Hello. Numbers. Colours. 1. This is me. 2. Clothes. 3. Family. 4. Home. 5. Food. 6.
 Contents Kuinka monta jakson sanaa opit? Väritä tähdet. Hello Numbers Colours 1. This is me 2. Clothes 3. Family 4. Home 5. Food 6. I can 7. School 8. Hobbies 9. Animals 10. Moving around EXTRA Hello Piirrä
Contents Kuinka monta jakson sanaa opit? Väritä tähdet. Hello Numbers Colours 1. This is me 2. Clothes 3. Family 4. Home 5. Food 6. I can 7. School 8. Hobbies 9. Animals 10. Moving around EXTRA Hello Piirrä
Tuote nro Tuotenimi Ovh Alv22 Tuoteryhmä Lyhyt nro
 Tuote nro Tuotenimi Ovh Alv22 Tuoteryhmä Lyhyt nro A544402 AFA + 544402 66,00 1000 A544402 A553400 AFA + 553400 70,00 1000 A553400 A556400 AFA + 556400 72,00 1000 A556400 A560412 AFA + 560412 84,00 1000
Tuote nro Tuotenimi Ovh Alv22 Tuoteryhmä Lyhyt nro A544402 AFA + 544402 66,00 1000 A544402 A553400 AFA + 553400 70,00 1000 A553400 A556400 AFA + 556400 72,00 1000 A556400 A560412 AFA + 560412 84,00 1000
KASVIKUNTA kl 2016
 526128 KASVIKUNTA kl 2016 Luennot klo 10-12 salissa 6602 11.4. Ma johdanto kasvikuntaan 12.4. Ti Chlorophyta 14.4. To Streptophyta 15.4. Pe vanhimmat alkiolliset kasvit 18.4. Ma maksasammalet 19.4. Ti
526128 KASVIKUNTA kl 2016 Luennot klo 10-12 salissa 6602 11.4. Ma johdanto kasvikuntaan 12.4. Ti Chlorophyta 14.4. To Streptophyta 15.4. Pe vanhimmat alkiolliset kasvit 18.4. Ma maksasammalet 19.4. Ti
Chapter 9 Motif finding. Chaochun Wei Spring 2019
 1896 1920 1987 2006 Chapter 9 Motif finding Chaochun Wei Spring 2019 Contents 1. Reading materials 2. Sequence structure modeling Motif finding Regulatory module finding 2 Reading materials Tompa et al
1896 1920 1987 2006 Chapter 9 Motif finding Chaochun Wei Spring 2019 Contents 1. Reading materials 2. Sequence structure modeling Motif finding Regulatory module finding 2 Reading materials Tompa et al
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.9.269
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.9.269
Supplementary Table S1. Genes that are upregulated during iron toxicity. Foldchange
 Supplementary Table S1. Genes that are upregulated during iron toxicity. Locus tag Spy Number a Foldchange b Annotation c Iron efflux pmta SpyM3_1093 17.6 Iron efflux pump Arginine and serine catabolism
Supplementary Table S1. Genes that are upregulated during iron toxicity. Locus tag Spy Number a Foldchange b Annotation c Iron efflux pmta SpyM3_1093 17.6 Iron efflux pump Arginine and serine catabolism
CS284A Representations & Algorithms for Molecular Biology. Xiaohui S. Xie University of California, Irvine
 CS284A Representations & Algorithms for Molecular Biology Xiaohui S. Xie University of California, Irvine Today s Goals Course information Challenges in computational biology Introduction to molecular
CS284A Representations & Algorithms for Molecular Biology Xiaohui S. Xie University of California, Irvine Today s Goals Course information Challenges in computational biology Introduction to molecular
Metsälamminkankaan tuulivoimapuiston osayleiskaava
 VAALAN KUNTA TUULISAIMAA OY Metsälamminkankaan tuulivoimapuiston osayleiskaava Liite 3. Varjostusmallinnus FCG SUUNNITTELU JA TEKNIIKKA OY 12.5.2015 P25370 SHADOW - Main Result Assumptions for shadow calculations
VAALAN KUNTA TUULISAIMAA OY Metsälamminkankaan tuulivoimapuiston osayleiskaava Liite 3. Varjostusmallinnus FCG SUUNNITTELU JA TEKNIIKKA OY 12.5.2015 P25370 SHADOW - Main Result Assumptions for shadow calculations
TM ETRS-TM35FIN-ETRS89 WTG
 VE1 SHADOW - Main Result Calculation: 8 x Nordex N131 x HH145m Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please
VE1 SHADOW - Main Result Calculation: 8 x Nordex N131 x HH145m Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please
Manolis Kellis Piotr Indyk
 6.095 / 6.895 Computational Biology: Genomes, Networks, Evolution Rapid database search Courtsey of CCRNP, The National Cancer Institute. Manolis Kellis Piotr Indyk Protein interaction network Courtesy
6.095 / 6.895 Computational Biology: Genomes, Networks, Evolution Rapid database search Courtsey of CCRNP, The National Cancer Institute. Manolis Kellis Piotr Indyk Protein interaction network Courtesy
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Calculation: N117 x 9 x HH141 Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG
SHADOW - Main Result Calculation: N117 x 9 x HH141 Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG
( ( OX2 Perkkiö. Rakennuskanta. Varjostus. 9 x N131 x HH145
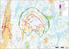 OX2 9 x N131 x HH145 Rakennuskanta Asuinrakennus Lomarakennus Liike- tai julkinen rakennus Teollinen rakennus Kirkko tai kirkollinen rak. Muu rakennus Allas Varjostus 1 h/a 8 h/a 20 h/a 0 0,5 1 1,5 2 km
OX2 9 x N131 x HH145 Rakennuskanta Asuinrakennus Lomarakennus Liike- tai julkinen rakennus Teollinen rakennus Kirkko tai kirkollinen rak. Muu rakennus Allas Varjostus 1 h/a 8 h/a 20 h/a 0 0,5 1 1,5 2 km
Tynnyrivaara, OX2 Tuulivoimahanke. ( Layout 9 x N131 x HH145. Rakennukset Asuinrakennus Lomarakennus 9 x N131 x HH145 Varjostus 1 h/a 8 h/a 20 h/a
 , Tuulivoimahanke Layout 9 x N131 x HH145 Rakennukset Asuinrakennus Lomarakennus 9 x N131 x HH145 Varjostus 1 h/a 8 h/a 20 h/a 0 0,5 1 1,5 km 2 SHADOW - Main Result Assumptions for shadow calculations
, Tuulivoimahanke Layout 9 x N131 x HH145 Rakennukset Asuinrakennus Lomarakennus 9 x N131 x HH145 Varjostus 1 h/a 8 h/a 20 h/a 0 0,5 1 1,5 km 2 SHADOW - Main Result Assumptions for shadow calculations
lpar1 IPB004065, IPB002277, and IPB Restriction Enyzme Differences from REBASE Gained in Variant Lost from Reference
 lpar1 IPB465, IPB2277, and IPB5385 Genomic Sequence Coding Sequence For help interpreting these results, view the PARSENP Introduction page. # View On Sequence Nucleotide Change Effect Restriction Enyzme
lpar1 IPB465, IPB2277, and IPB5385 Genomic Sequence Coding Sequence For help interpreting these results, view the PARSENP Introduction page. # View On Sequence Nucleotide Change Effect Restriction Enyzme
Genomic and epigenomic signatures for interpreting complex disease
 Genomic and epigenomic signatures for interpreting complex disease Manolis Kellis Broad Institute of MIT and Harvard MIT Computer Science & Artificial Intelligence Laboratory TATTGAATTTTCAAAAATTCTTACTTTTTTTTTGGATGGACGCAAAGAAGTTTAATAATCATATTACATGGCATTACCACCATATA
Genomic and epigenomic signatures for interpreting complex disease Manolis Kellis Broad Institute of MIT and Harvard MIT Computer Science & Artificial Intelligence Laboratory TATTGAATTTTCAAAAATTCTTACTTTTTTTTTGGATGGACGCAAAGAAGTTTAATAATCATATTACATGGCATTACCACCATATA
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
,0 Yes ,0 120, ,8
 SHADOW - Main Result Calculation: Alue 2 ( x 9 x HH120) TuuliSaimaa kaavaluonnos Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered
SHADOW - Main Result Calculation: Alue 2 ( x 9 x HH120) TuuliSaimaa kaavaluonnos Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered
Supplementary Information
 Supplementary Information Quantifying Absolute Addressability in DNA origami with Molecular Resolution Maximilian T. Strauss, Florian Schueder, Daniel Haas, Philipp C. Nickels, and Ralf Jungmann Supplementary
Supplementary Information Quantifying Absolute Addressability in DNA origami with Molecular Resolution Maximilian T. Strauss, Florian Schueder, Daniel Haas, Philipp C. Nickels, and Ralf Jungmann Supplementary
Petopia Imports Oy Toukolantie 19, Pertteli puh. (02) matkap fax (02)
 Classics koiran pannat Lila DC-ZZ-PU-SM DC-ZZ-OR-SM DC-ZZ-PU-ME DC-ZZ-OR-ME DC-ZZ-PU-LG 25mm 40-60cm DC-ZZ-OR-LG 25mm 40-60cm Vaaleanpunainen Vaaleansininen DC-ZZ-PK-SM DC-ZZ-LB-SM DC-ZZ-PK-ME DC-ZZ-LB-ME
Classics koiran pannat Lila DC-ZZ-PU-SM DC-ZZ-OR-SM DC-ZZ-PU-ME DC-ZZ-OR-ME DC-ZZ-PU-LG 25mm 40-60cm DC-ZZ-OR-LG 25mm 40-60cm Vaaleanpunainen Vaaleansininen DC-ZZ-PK-SM DC-ZZ-LB-SM DC-ZZ-PK-ME DC-ZZ-LB-ME
Lampiran 1 Komposisi media pertumbuhan yang digunakan A. Media nutrien agar (NA) - Agar 1.5% - Nutrient broth 0.8%
 LAMPIRAN 8 9 Lampiran 1 Komposisi media pertumbuhan yang digunakan A. Media nutrien agar (NA) - Agar 1.5% - Nutrient broth 0.8% B. Media King s B agar - Agar 1.5% - Pepton 2% - K 2 HPO 4 0.15% - MgSO 4.4H
LAMPIRAN 8 9 Lampiran 1 Komposisi media pertumbuhan yang digunakan A. Media nutrien agar (NA) - Agar 1.5% - Nutrient broth 0.8% B. Media King s B agar - Agar 1.5% - Pepton 2% - K 2 HPO 4 0.15% - MgSO 4.4H
Säteilyturvakeskuksen määräys turvallisuusluvasta ja valvonnasta vapauttamisesta
 1 (33) LUONNOS 2 -MÄÄRÄYS STUK SY/1/2017 Säteilyturvakeskuksen määräys turvallisuusluvasta ja valvonnasta vapauttamisesta Säteilyturvakeskuksen päätöksen mukaisesti määrätään säteilylain ( / ) 49 :n 3
1 (33) LUONNOS 2 -MÄÄRÄYS STUK SY/1/2017 Säteilyturvakeskuksen määräys turvallisuusluvasta ja valvonnasta vapauttamisesta Säteilyturvakeskuksen päätöksen mukaisesti määrätään säteilylain ( / ) 49 :n 3
Date Päiväys J.Mikkonen Signature Allekirjoitus. V.Tepponen
 Changes in the drawing are allowed only by the permission of the authorities who have granted the certificate Muutokset sallittu vain sertifikaatin myöntäjän luvalla The drawing is a valid document only
Changes in the drawing are allowed only by the permission of the authorities who have granted the certificate Muutokset sallittu vain sertifikaatin myöntäjän luvalla The drawing is a valid document only
SUODATIN- PATRUUNAT MASINO-HYDROSTO KEY OY
 1 SUODATIN- PATRUUNAT 2006 10 MASINO- KEY OY 2 Masino-Hydrosto key Oy toimittaa suodatusjärjestelmiä, suodattimia ja patruunoita hydrauli- ja kiertovoitelujärjestelmiin, kompressoreihin, ilmalle, vedelle
1 SUODATIN- PATRUUNAT 2006 10 MASINO- KEY OY 2 Masino-Hydrosto key Oy toimittaa suodatusjärjestelmiä, suodattimia ja patruunoita hydrauli- ja kiertovoitelujärjestelmiin, kompressoreihin, ilmalle, vedelle
WindPRO version joulu 2012 Printed/Page :42 / 1. SHADOW - Main Result
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table 13.6.2013 19:42 / 1 Minimum
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table 13.6.2013 19:42 / 1 Minimum
MCV 1,5/ 3-G-3,81. Poimi online-katalogista. Tilausnumero:
 Poimi online-katalogista MCV 1,5/ 3-G-3,81 Tilausnumero: 1803439 The figure shows a 10-position version of the product Header, Nominal current:, Nom. voltage: 160 V, Pitch: 3.81 mm, Number of positions:
Poimi online-katalogista MCV 1,5/ 3-G-3,81 Tilausnumero: 1803439 The figure shows a 10-position version of the product Header, Nominal current:, Nom. voltage: 160 V, Pitch: 3.81 mm, Number of positions:
3 *ä;r ä:e 5ä ä{ :i. c oo) S g+;!qg *r; Er ; l[$ E ;;iä F:ä ä :E ä: a bo. =. * gäf$iery g! Eä. a is äg*!=."fl: ä; E!, \ ins:" qgg ;._ EE üg.
 t AJ 1., t4 t4 \J : h J \) (.) \ ( J r ) tḡr (u (1) m * t *h& r( t{ L.C g :LA( g9; p ö m. gr iop ö O t : U 0J (U.p JJ! ä; >
t AJ 1., t4 t4 \J : h J \) (.) \ ( J r ) tḡr (u (1) m * t *h& r( t{ L.C g :LA( g9; p ö m. gr iop ö O t : U 0J (U.p JJ! ä; >
( ,5 1 1,5 2 km
 Tuulivoimala Rakennukset Asuinrakennus Liikerak. tai Julkinen rak. Lomarakennus Teollinen rakennus Kirkollinen rakennus Varjostus "real case" h/a 1 h/a 8 h/a 20 h/a 4 5 3 1 2 6 7 8 9 10 0 0,5 1 1,5 2 km
Tuulivoimala Rakennukset Asuinrakennus Liikerak. tai Julkinen rak. Lomarakennus Teollinen rakennus Kirkollinen rakennus Varjostus "real case" h/a 1 h/a 8 h/a 20 h/a 4 5 3 1 2 6 7 8 9 10 0 0,5 1 1,5 2 km
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table 22.12.2014 11:33 / 1 Minimum
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table 22.12.2014 11:33 / 1 Minimum
WindPRO version joulu 2012 Printed/Page :47 / 1. SHADOW - Main Result
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
LVI-Dahl Oy Robert Huberin tie 5, PL 212, Vantaa puh , faksi ,
 Painemittarihanat S004.16. Painemittarihanat DIN 16270 Materiaalit: Messinki, 1.0460, 1.4571 Messinki C.22.8 1.4571 120 C 250 bar 400 bar 400 bar 200 C - 300 bar 280 bar 16270A 250-10 +120 100 79 63 27
Painemittarihanat S004.16. Painemittarihanat DIN 16270 Materiaalit: Messinki, 1.0460, 1.4571 Messinki C.22.8 1.4571 120 C 250 bar 400 bar 400 bar 200 C - 300 bar 280 bar 16270A 250-10 +120 100 79 63 27
MC 1,5/10-G-3,81. Poimi online-katalogista. Tilausnumero:
 Poimi online-katalogista MC 1,5/10-G-3,81 Tilausnumero: 1803358 The figure shows a 10-position version of the product Header, Nominal current:, Nom. voltage: 160 V, Pitch: 3.81 mm, Number of positions:
Poimi online-katalogista MC 1,5/10-G-3,81 Tilausnumero: 1803358 The figure shows a 10-position version of the product Header, Nominal current:, Nom. voltage: 160 V, Pitch: 3.81 mm, Number of positions:
In situ hybridization
 In situ hybridization ISH Detection of DNA or RNA Single or double stranded Chromosomal or cellular nucleic acids ISH Type of a hybrid? DNA-DNA In situ renaturation of target DNA in ISH cannot be prevented
In situ hybridization ISH Detection of DNA or RNA Single or double stranded Chromosomal or cellular nucleic acids ISH Type of a hybrid? DNA-DNA In situ renaturation of target DNA in ISH cannot be prevented
Rekisteröiminen - FAQ
 Rekisteröiminen - FAQ Miten Akun/laturin rekisteröiminen tehdään Akun/laturin rekisteröiminen tapahtuu samalla tavalla kuin nykyinen takuurekisteröityminen koneille. Nykyistä tietokantaa on muokattu niin,
Rekisteröiminen - FAQ Miten Akun/laturin rekisteröiminen tehdään Akun/laturin rekisteröiminen tapahtuu samalla tavalla kuin nykyinen takuurekisteröityminen koneille. Nykyistä tietokantaa on muokattu niin,
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table 5.11.2013 16:44 / 1 Minimum
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table 5.11.2013 16:44 / 1 Minimum
SUOMEN SALIBANDYLIITTO r.y turnaus,kierrokset 1-2 Erotuomariasettelija Tero Merenheimo
 1.turnaus,kierrokset 1-2 Erotuomariasettelija Tero Merenheimo 19.11.2016 09:00 PC2-A ES Oilers Sininen Indians Esport Areena 4 Espoo Oilers Sininen 19.11.2016 10:00 PC2-A ES SB Vantaa Black Panthers Esport
1.turnaus,kierrokset 1-2 Erotuomariasettelija Tero Merenheimo 19.11.2016 09:00 PC2-A ES Oilers Sininen Indians Esport Areena 4 Espoo Oilers Sininen 19.11.2016 10:00 PC2-A ES SB Vantaa Black Panthers Esport
Changes in the drawing are allowed only by the permission of the authorities who have granted the certificate Muutokset sallittu vain sertifikaatin my
 Changes in the drawing are allowed only by the permission of the authorities who have granted the certificate Muutokset sallittu vain sertifikaatin myöntäjän luvalla The drawing is a valid document only
Changes in the drawing are allowed only by the permission of the authorities who have granted the certificate Muutokset sallittu vain sertifikaatin myöntäjän luvalla The drawing is a valid document only
ELEMET- MOCASTRO. Effect of grain size on A 3 temperatures in C-Mn and low alloyed steels - Gleeble tests and predictions. Period
 1 ELEMET- MOCASTRO Effect of grain size on A 3 temperatures in C-Mn and low alloyed steels - Gleeble tests and predictions Period 20.02-25.05.2012 Diaarinumero Rahoituspäätöksen numero 1114/31/2010 502/10
1 ELEMET- MOCASTRO Effect of grain size on A 3 temperatures in C-Mn and low alloyed steels - Gleeble tests and predictions Period 20.02-25.05.2012 Diaarinumero Rahoituspäätöksen numero 1114/31/2010 502/10
Rakennukset Varjostus "real case" h/a 0,5 1,5
 Tuulivoimala Rakennukset Asuinrakennus Liikerak. tai Julkinen rak. Lomarakennus Teollinen rakennus Kirkollinen rakennus Varjostus "real case" h/a 1 h/a 8 h/a 20 h/a 1 2 3 5 8 4 6 7 9 10 0 0,5 1 1,5 2 km
Tuulivoimala Rakennukset Asuinrakennus Liikerak. tai Julkinen rak. Lomarakennus Teollinen rakennus Kirkollinen rakennus Varjostus "real case" h/a 1 h/a 8 h/a 20 h/a 1 2 3 5 8 4 6 7 9 10 0 0,5 1 1,5 2 km
State of the Union... Functional Genomics Research Stream. Molecular Biology. Genomics. Computational Biology
 Functional Genomics Research Stream State of the Union... Research Meeting: February 16, 2010 Functional Genomics & Research Report III Concepts Genomics Molecular Biology Computational Biology Genome
Functional Genomics Research Stream State of the Union... Research Meeting: February 16, 2010 Functional Genomics & Research Report III Concepts Genomics Molecular Biology Computational Biology Genome
LUONNOS RT 80260 EN AGREEMENT ON BUILDING WORKS 1 THE PARTIES. May 1998 1 (10)
 RT 80260 EN May 1998 1 (10) AGREEMENT ON BUILDING WORKS This agreement template is based on the General Terms and Conditions of Building Contracts YSE 1998 RT 16-10660, LVI 03-10277, Ratu 417-7, KH X4-00241.
RT 80260 EN May 1998 1 (10) AGREEMENT ON BUILDING WORKS This agreement template is based on the General Terms and Conditions of Building Contracts YSE 1998 RT 16-10660, LVI 03-10277, Ratu 417-7, KH X4-00241.
Genome 373: Genomic Informatics. Professors Elhanan Borenstein and Jay Shendure
 Genome 373: Genomic Informatics Professors Elhanan Borenstein and Jay Shendure Genome 373 This course is intended to introduce students to the breadth of problems and methods in computational analysis
Genome 373: Genomic Informatics Professors Elhanan Borenstein and Jay Shendure Genome 373 This course is intended to introduce students to the breadth of problems and methods in computational analysis
SUOMEN SALIBANDYLIITTO r.y Maaniitun koulu Heikkiläntie Nurmijärvi 1.turnaus,kierrokset 1-2 Erotuomariasettelija Tero Merenheimo
 Maaniitun koulu Heikkiläntie 27 01900 Nurmijärvi 1.turnaus,kierrokset 1-2 Erotuomariasettelija Tero Merenheimo 18.11.2017 09:00 TB-A ES SB-Pro Northern Stars Maaniitun koulu 1 Nurmijärvi SB-Pro 18.11.2017
Maaniitun koulu Heikkiläntie 27 01900 Nurmijärvi 1.turnaus,kierrokset 1-2 Erotuomariasettelija Tero Merenheimo 18.11.2017 09:00 TB-A ES SB-Pro Northern Stars Maaniitun koulu 1 Nurmijärvi SB-Pro 18.11.2017
( N117 x HH141 ( Honkajoki N117 x 9 x HH120 tv-alueet ( ( ( ( ( ( ( ( ( ( m. Honkajoki & Kankaanpää tuulivoimahankkeet
 Honkajoki & Kankaanpää tuulivoimahankkeet N117 x HH141 Honkajoki N117 x 9 x HH120 tv-alueet Alahonkajoki_kaava_alueen_raja_polyline Asuinrakennus Julkinen tai liiker rak. Lomarakennus Teollinen rak. Allas
Honkajoki & Kankaanpää tuulivoimahankkeet N117 x HH141 Honkajoki N117 x 9 x HH120 tv-alueet Alahonkajoki_kaava_alueen_raja_polyline Asuinrakennus Julkinen tai liiker rak. Lomarakennus Teollinen rak. Allas
Eo C)sl. oarl. d to E= J. o-= o cy) =uo. f,e. ic v. .o6. .9o. äji. :ir. ijo 96. {c o o. ';i _o. :fe. C=?i. t-l +) (- c rt, u0 C.
 C C C)l A\ d Y) L P C v J J rl, ( 0 C.6 +) ( j 96.9 :r : C (Db]? d '; _ äj r, { . 3 k l: d d 6 60QOO:ddO 96.l ä.c p _ : 6 äp l P C..86 p r5 r!l (, ō J. J rl r O 6!6 (5 ) ä dl r l { ::: :: :: 6e g r : ;
C C C)l A\ d Y) L P C v J J rl, ( 0 C.6 +) ( j 96.9 :r : C (Db]? d '; _ äj r, { . 3 k l: d d 6 60QOO:ddO 96.l ä.c p _ : 6 äp l P C..86 p r5 r!l (, ō J. J rl r O 6!6 (5 ) ä dl r l { ::: :: :: 6e g r : ;
Asennusohjeet P/N MMI , Rev. A Syyskuu ATEX -asennusohjeet Micro Motion MVD Direct Connect -mittareille
 Asennusohjeet P/N MMI-20011744, Rev. A Syyskuu 2008 ATEX -asennusohjeet Micro Motion MVD Direct Connect -mittareille Huomautus: kun kyseessä ovat vaaralliset asennukset Euroopassa, katso standardia EN
Asennusohjeet P/N MMI-20011744, Rev. A Syyskuu 2008 ATEX -asennusohjeet Micro Motion MVD Direct Connect -mittareille Huomautus: kun kyseessä ovat vaaralliset asennukset Euroopassa, katso standardia EN
Title: Enhancement of protein production via the strong DIT1 terminator and two RNA-binding proteins in Saccharomyces cerevisiae
 Title: Enhancement of protein production via the strong DIT1 terminator and two RNA-binding proteins in Saccharomyces cerevisiae Authors: Yoichiro Ito, Takao Kitagawa, Mamoru Yamanishi, Satoshi Katahira,
Title: Enhancement of protein production via the strong DIT1 terminator and two RNA-binding proteins in Saccharomyces cerevisiae Authors: Yoichiro Ito, Takao Kitagawa, Mamoru Yamanishi, Satoshi Katahira,
Asennusohjeet P/N MMI , Rev. A Heinäku ATEX-asennusohjeet Micro Motion -mallien 1500 ja 2500 lähettimille
 Asennusohjeet P/N MMI-20011739, Rev. A Heinäku 2008 ATEX-asennusohjeet Micro Motion -mallien 1500 ja 2500 lähettimille Huomautus: kun kyseessä ovat vaaralliset asennukset Euroopassa, katso standardia EN
Asennusohjeet P/N MMI-20011739, Rev. A Heinäku 2008 ATEX-asennusohjeet Micro Motion -mallien 1500 ja 2500 lähettimille Huomautus: kun kyseessä ovat vaaralliset asennukset Euroopassa, katso standardia EN
SIDEAINE: Alkydisidos, vesipohjainen
 PAINTS GENERAL PURPOSE NEW SECURESEAL SECURE C A P S E A L E D ZERO SPRAYPAINT THE 1ST SOLVENT FREE SPRAYPAINT IN THE WORLD 3m 2 (400 ml) COVERAGE MATERIAL HARDNESS NO SMELL USE PLASTIC PRIMER EASY CLEAN
PAINTS GENERAL PURPOSE NEW SECURESEAL SECURE C A P S E A L E D ZERO SPRAYPAINT THE 1ST SOLVENT FREE SPRAYPAINT IN THE WORLD 3m 2 (400 ml) COVERAGE MATERIAL HARDNESS NO SMELL USE PLASTIC PRIMER EASY CLEAN
Changes in the drawing are allowed only by the permission of the authorities who have granted the certificate Muutokset sallittu vain sertifikaatin my
 Changes in the drawing are allowed only by the permission of the authorities who have granted the certificate Muutokset sallittu vain sertifikaatin myöntäjän luvalla The drawing is a valid document only
Changes in the drawing are allowed only by the permission of the authorities who have granted the certificate Muutokset sallittu vain sertifikaatin myöntäjän luvalla The drawing is a valid document only
Supplementary Information
 Supplementary Information Supplementary Table 1. Strain table Strain name Genotype Reference CEN.PK111-27B MATa leu2 trp1 Euroscarf TC-49 TC-50 pdc5::aro4* aro10::aro7* This study TC-50 MATa leu2 trp1
Supplementary Information Supplementary Table 1. Strain table Strain name Genotype Reference CEN.PK111-27B MATa leu2 trp1 Euroscarf TC-49 TC-50 pdc5::aro4* aro10::aro7* This study TC-50 MATa leu2 trp1
Figure S1. CLUSTAL O(1.2.1) multiple sequence alignment
 Figure S1. A CLUSTAL O(1.2.1) multiple sequence alignment AT5G48657.1 --------------------------------MNVANPNEYPDMNPNAAQNRNMSRPDQQ 28 AT5G48657.2 MANRPHVPKFGDWTEDAPFTVVFEKASKSKKNMNVANPNEYPDMNPNAAQNRNMSRPDQQ
Figure S1. A CLUSTAL O(1.2.1) multiple sequence alignment AT5G48657.1 --------------------------------MNVANPNEYPDMNPNAAQNRNMSRPDQQ 28 AT5G48657.2 MANRPHVPKFGDWTEDAPFTVVFEKASKSKKNMNVANPNEYPDMNPNAAQNRNMSRPDQQ
Table S2. S. thermophilus CRISPR spacer sequence analysis.
 Table S2. S. thermophilus CRISPR analysis. 001_1_01 TGTTTGACAGCAAATCAAGATTCGAATTGT 30 AF115103 Streptococcus thermophilus bacteriophage Sfi21, compl 40 93 28 2 0 3 30 38304 38331 1 001_1_02 AATGACGAGGAGCTATTGGCACAACTTACA
Table S2. S. thermophilus CRISPR analysis. 001_1_01 TGTTTGACAGCAAATCAAGATTCGAATTGT 30 AF115103 Streptococcus thermophilus bacteriophage Sfi21, compl 40 93 28 2 0 3 30 38304 38331 1 001_1_02 AATGACGAGGAGCTATTGGCACAACTTACA
Supporting Information Table S1 Core regions of 18 identified promoters in C. acetobutylicum
 Supporting Information Table S1 Core regions of 18 identified promoters in C. acetobutylicum Gene Core regions of the promoters Reference 35 region 10 region thl TATA TTGATA AAAATAATAATAGTGGG TATAAT TAA
Supporting Information Table S1 Core regions of 18 identified promoters in C. acetobutylicum Gene Core regions of the promoters Reference 35 region 10 region thl TATA TTGATA AAAATAATAATAGTGGG TATAAT TAA
SUOMEN SALIBANDYLIITTO r.y turnaus,kierrokset Erotuomariasettelija Tero Merenheimo
 F2-POJAT HAASTAJASARJA ALEMPI, I-RYHMÄ 10.03.2019 09:00 PF2-I ES FC Tuusula SB-Pro Tuusulan SBH 1 Tuusula FC Tuusula 10.03.2019 09:45 PF2-I ES AskU SC Hawks Yellow Tuusulan SBH 1 Tuusula FC Tuusula 10.03.2019
F2-POJAT HAASTAJASARJA ALEMPI, I-RYHMÄ 10.03.2019 09:00 PF2-I ES FC Tuusula SB-Pro Tuusulan SBH 1 Tuusula FC Tuusula 10.03.2019 09:45 PF2-I ES AskU SC Hawks Yellow Tuusulan SBH 1 Tuusula FC Tuusula 10.03.2019
Hiukkaskoko maks. 5 µm. Mäntään kohdistuvan voiman mittapaine 6,3 bar. Materiaalit:
 1 Käyttöpaine min./max. 2 bar / 8 bar Ympäristölämpötila min./maks. -10 C / +60 C Keski Paineilma Hiukkaskoko maks. 5 µm Paineilman öljypitoisuus 0 mg/m³ - 1 mg/m³ Mäntään kohdistuvan voiman mittapaine
1 Käyttöpaine min./max. 2 bar / 8 bar Ympäristölämpötila min./maks. -10 C / +60 C Keski Paineilma Hiukkaskoko maks. 5 µm Paineilman öljypitoisuus 0 mg/m³ - 1 mg/m³ Mäntään kohdistuvan voiman mittapaine
Capacity Utilization
 Capacity Utilization Tim Schöneberg 28th November Agenda Introduction Fixed and variable input ressources Technical capacity utilization Price based capacity utilization measure Long run and short run
Capacity Utilization Tim Schöneberg 28th November Agenda Introduction Fixed and variable input ressources Technical capacity utilization Price based capacity utilization measure Long run and short run
Bioinformatics. Sequence Analysis: Part III. Pattern Searching and Gene Finding. Fran Lewitter, Ph.D. Head, Biocomputing Whitehead Institute
 Bioinformatics Sequence Analysis: Part III. Pattern Searching and Gene Finding Fran Lewitter, Ph.D. Head, Biocomputing Whitehead Institute Course Syllabus Jan 7 Jan 14 Jan 21 Jan 28 Feb 4 Feb 11 Feb 18
Bioinformatics Sequence Analysis: Part III. Pattern Searching and Gene Finding Fran Lewitter, Ph.D. Head, Biocomputing Whitehead Institute Course Syllabus Jan 7 Jan 14 Jan 21 Jan 28 Feb 4 Feb 11 Feb 18
KONEISTUSKOKOONPANON TEKEMINEN NX10-YMPÄRISTÖSSÄ
 KONEISTUSKOKOONPANON TEKEMINEN NX10-YMPÄRISTÖSSÄ https://community.plm.automation.siemens.com/t5/tech-tips- Knowledge-Base-NX/How-to-simulate-any-G-code-file-in-NX- CAM/ta-p/3340 Koneistusympäristön määrittely
KONEISTUSKOKOONPANON TEKEMINEN NX10-YMPÄRISTÖSSÄ https://community.plm.automation.siemens.com/t5/tech-tips- Knowledge-Base-NX/How-to-simulate-any-G-code-file-in-NX- CAM/ta-p/3340 Koneistusympäristön määrittely
SUORITUSTASOILMOITUS Nro: DoP [FI]
![SUORITUSTASOILMOITUS Nro: DoP [FI] SUORITUSTASOILMOITUS Nro: DoP [FI]](/thumbs/70/62877789.jpg) Tuotetyypin yksilöllinen tunniste: ESSVE ESSD (self-drilling) fastening screws ESSVE ESST (self-tapping) fastening screws Aiottu käyttötarkoitus (aiotut käyttötarkoitukset): The fastening screws are intended
Tuotetyypin yksilöllinen tunniste: ESSVE ESSD (self-drilling) fastening screws ESSVE ESST (self-tapping) fastening screws Aiottu käyttötarkoitus (aiotut käyttötarkoitukset): The fastening screws are intended
T Statistical Natural Language Processing Answers 6 Collocations Version 1.0
 T-61.5020 Statistical Natural Language Processing Answers 6 Collocations Version 1.0 1. Let s start by calculating the results for pair valkoinen, talo manually: Frequency: Bigrams valkoinen, talo occurred
T-61.5020 Statistical Natural Language Processing Answers 6 Collocations Version 1.0 1. Let s start by calculating the results for pair valkoinen, talo manually: Frequency: Bigrams valkoinen, talo occurred
DS-tunnusten haku Outi Jäppinen CIMO
 DS-tunnusten haku 2013 Outi Jäppinen CIMO 2/2009 DS-tunnukset ECTS- ja DS-tunnusten avulla pyritään edistämään ECTS-järjestelmän sekä tutkintotodistuksen liitteen Diploma Supplementin asianmukaista käyttöä
DS-tunnusten haku 2013 Outi Jäppinen CIMO 2/2009 DS-tunnukset ECTS- ja DS-tunnusten avulla pyritään edistämään ECTS-järjestelmän sekä tutkintotodistuksen liitteen Diploma Supplementin asianmukaista käyttöä
1 3StoVentec FI - j 0 1rjestelm 0 1kuvaus_200320133
 g 0 3 ē 0 7 0 5 Ē 0 7 0 1 0 0 0 3 0 9 Ŧ 0 6~ Ŧ 0 5 p Ē 0 4s 0 7 0 2 Ē 0 7 0 3g Ŧ 0 1 0 9 ĸ 0 5 ĸ 0 6 Ŧ 0 3 ē 0 7 0 5 Ē 0 7 0 1 0 0 0 3 ē~ 0 2 0 9~ ē~ 0 9 0 7 0 2 Ē 0 7 0 7 0 1 0 1 0 7 v 0 1 0 7 ĸ 0 5 Ē
g 0 3 ē 0 7 0 5 Ē 0 7 0 1 0 0 0 3 0 9 Ŧ 0 6~ Ŧ 0 5 p Ē 0 4s 0 7 0 2 Ē 0 7 0 3g Ŧ 0 1 0 9 ĸ 0 5 ĸ 0 6 Ŧ 0 3 ē 0 7 0 5 Ē 0 7 0 1 0 0 0 3 ē~ 0 2 0 9~ ē~ 0 9 0 7 0 2 Ē 0 7 0 7 0 1 0 1 0 7 v 0 1 0 7 ĸ 0 5 Ē
Koppisiemeniset eli kukkakasvit. Koppisiemeniset: Yleiset piirteet Kasvisystematiikka: maakasvit. Sukupolvenvuorottelu. Koppisiemenisten ekspansio
 eli kukkakasvit Yleiskuva : Yleiset piirteet Kasvisystematiikka: maakasvit Jari Oksanen Biologian tutkinto-ohjelma 2016 Viridiplantae muut viherlevät sisarlevät sammalet jne maakasvit lieot putkilokasvit
eli kukkakasvit Yleiskuva : Yleiset piirteet Kasvisystematiikka: maakasvit Jari Oksanen Biologian tutkinto-ohjelma 2016 Viridiplantae muut viherlevät sisarlevät sammalet jne maakasvit lieot putkilokasvit
FYSE301(Elektroniikka(1(A3osa,(kevät(2013(
 FYSE301(Elektroniikka(1(A3osa,(kevät(2013( 1/2 Loppukoe1.3.2013 vastaakaikkiinkysymyksiin(yhteensä48pistettä) 1. Kuvailelyhyesti a. Energialineaarisissapiirielementeissä:vastuksessa,kondensaattorissajakelassa(3
FYSE301(Elektroniikka(1(A3osa,(kevät(2013( 1/2 Loppukoe1.3.2013 vastaakaikkiinkysymyksiin(yhteensä48pistettä) 1. Kuvailelyhyesti a. Energialineaarisissapiirielementeissä:vastuksessa,kondensaattorissajakelassa(3

