The RPCI-1, 3, 4, and 5 human P1 artificial chromosome (PAC), the RPCI-11(Ref) 1
|
|
|
- Aarne Turunen
- 5 vuotta sitten
- Katselukertoja:
Transkriptio
1 Supplementary Information A. Genomics The RPCI-1, 3, 4, and 5 human P1 artificial chromosome (PAC), the RPCI-11(Ref) 1 human, the RPCI-23(Ref) 2 mouse (B6) and the DIL-NOD bacterial artificial chromosome (BAC) libraries were obtained (Table A1). For all RPCI library information see The DIL-NOD library was prepared from the NOD/MrkTac mouse at the Institute of Physical and Chemical Research (RIKEN Ibaraki, Japan) and is available for distribution from the JDRF/WT DIL. A number of probes were used to screen the libraries (data not shown). From the resulting positive clones six were sequenced (Table A1). Table A1: Library screening and sequencing information. Library Species/str Probe Sequenced Accession number ain clone name RPCI-1 Human (AT) n 3 UTR 61E2 AF RPCI-11 Human (AT) n 3 UTR 278L16 AJ535716, AJ535717, AJ RPCI-1 Human CD28 219D7 AF RPCI-23 Mouse B6 Cd28 157N11 AL RPCI-23 Mouse B6 Ctla4 146J17 AL DIL-NOD Mouse NOD Ctla4/Icos 257N2 AL The human and mouse sequences were assembled and used in database searches. The homology information was integrated into T1Dace, our in-house 1
2 adaptation of ACeDB 3 with the sequence, repeat information (RepeatMasker A.F.A. Smit and P. Green, unpublished, automatically annotated genes from EnsEMBL ( primers used for SNP harvesting, the identified SNPs, and SNPs from the public domain (dbsnp, TSC SNPs and HGBASE). The mouse and human genomic sequences were compared by constructing percent-identity plots (PIP) 4 (Figure A1). A view of this region using Generic Genome Browser (Gbrowse) ( is provided (Figure. A2). Only three functional genes exist in the region, CD28, CTLA4 and ICOS, and three pseudogenes (a sequence with similarity to the ubiquinoneoxidoreductase MLRQ subunit gene, keratin, and nucleolar phosphoprotein). A sequence alignment of mouse and human sequence around the CT60 SNP revealed 66.1% homology from the poly-a signal to CT60 (ClustalW, 1.74, (Figure A3). Fig. A1 2
3 Figure A1 3
4 4
5 5
6 6
7 7
8 human mouse +1772(polyA) AACTCAATATTTTCCATGAAAATGCAACAACATGTATAATATTTTTAATTAAATAAAAAT AACTCAATATTTTCCATGAAAATGCAACCACATGTATAATATTTTTAATTAAATAAAAGT **************************** ***************************** * human mouse human mouse CTGTGGTGGTCGTTTTCCGGAGTTGTC--TTTATCATCCTTGCATTTGAATATTGTGTTT TTCTTGTGATTGTTTTATGGGCTGCCCAATTTATCAGCTTTGTTTTTAAGTGTAGTGTTT * * *** * ***** ** * * ******* * *** *** * * * ****** AAAT TTTTGATTGATTCATTC GTGTTTGGTTTTGAAAAATGTTTTCAATTACGTGTGCATTTGTTTTTTATGGATTCATTC **** **** ** ********* human mouse human mouse human mouse human mouse human mouse AGTATCTGGTGGAGTCTCCAATATTAGAAATACTGGAAACAAACTGAAAAACCACAAAAG AGCACTAAGTAGAGCCTTTTATGCTAAAAATCCTGGAAACAAACTGGATCAGTGAAAAGA ** * ** *** ** ** ** **** ************** * * *** GACAAATAATGCTTCATGAGTCAGCTTTGCACCAGCCATTACCTGCAAGTCATTCTTGGA GGAAAAGAATATTCTATGGATCAGCCATGTACCAGCCGTTATCTGAAGCCCATTCCTGGA * *** *** * *** ***** ** ******* *** *** * ***** **** CT60 AGGTATCCATCCTCTTTCCTTTTGATTTCTTCACCACTATTTGGGATATAACGTGGGTTA AAGAGTCTATCATGT--ACTCTTAATTTCTT-----CTTTTTGG---ATAA A * * ** *** * * ** ** ******* ** ***** **** * ACACAGACATAGCAGTCCTTTATAAATCAATTGGCATGCTGTTTAACACAGGTTCTTCAC ATTCAGGCACAACCATCCTTTAAACTTTACACGCAGTTCTCTTTATC-CAGATTCCCCAC * *** ** * * ******* * * * * * ** **** * *** *** *** CTCCCCTTTCTTACC TTCCTCTTTCTTACC *** ********** Figure A3. Pairwise alignment of the human and mouse CT60 region. Asterisks show identities between the two sequences and dashes denote gaps in the alignment. A polyadenylation site in the human sequence is shown at position +1,772 with respect to the stop codon of CTLA4. The symbol marks the position at which a B3A SINE element was deleted from the mouse sequence to enable a better alignment between the two species. There is no corresponding element in the human sequence. The conservation in the 3 UTR of CTLA4 may suggest a functional role for this region. 8
9 SNP identification The scanned region and the three SNP identification methods used, are shown in Figure A2. PCR products from 32 individuals (the parents of eight probands from UK T1D families, and eight male and eight female control subjects) were scanned, using denaturing HPLC (Transgenomic Inc., Santa Clara, CA). Primers sets (136 in total) were designed for PCR products ( bp) using Primer3 ( and amplified in each of the 32 individuals and the products analysed according to Transgenomic Application Note 101. Samples with heteroduplex or different homoduplex patterns were then directly sequenced using the same PCR primers (Method 1). Direct sequencing of PCR products from 32 individuals was performed using an Applied Biosystems (ABI) 3700 capillary sequencer (Foster City, CA)(Method 2). For methods 1 and 2, the sequences from AF and RPCI L16 were used for SNP harvesting. SNPs from the public database (dbsnp) were extracted (Method 3) (Table A2). In total, more than 60% percent of the 317 kb sequence was scanned in 32 individuals for SNPs (Figure A2), excluding repeats sequences. All 28.6 kb of the CTLA-4 gene and its flanking DNA from 3.5 kb upstream of 5 UTR to 19 kb downstream of the published 3 UTR was contiguously scanned/re-sequenced. For CD kb was sequenced. This corresponds to 2.1 kb upstream of 5 UTR, all four exons, and 2.6 kb downstream of 3 UTR. For ICOS, SNPs were harvested from 13.9 kb corresponding to 2.7 kb upstream of the 5 UTR, the 5 exons, and all 3 UTR sequence. All the identified SNPs were mapped using the golden path sequence (NCBI build 28) and the information stored in our database. SNP coverage, and SNP positions are shown (Figure A2). Attempts were made to produce a denser SNP map between positions 100 kb and 150 kb using the public databases. No additional SNPs 9
10 were found. This may be due to the repetitive nature of this region. In total 108 SNPs were identified and/or confirmed. All the supporting information, including SNP sequences, shown in Table A3 is also available from our ftp site (ftp://ftp-gene.cimr.cam.ac.uk/ctla4). Table A2: The names of re-sequenced SNPs in dbsnp dbsnp identifiers confirmed/genotyped rs Yes rs Yes rs rs Yes rs rs rs Yes rs rs rs rs rs rs rs rs rs rs rs rs rs Yes Table A3: Sequences of 108 SNPs SNP names 1CD28p5_1Rb 2rs rs rs CD28ex3F 6rs rs V 8rs V 9CTAF185 10CTAF212 11CTAF305 SNP sequences AATATTAGCAGTGTTTATATTAGGGTG[T/G]TAAGAATGTGGATGAATCTTGTTTTAA T CTGGTGGAATAACCCTCTCT[T/G]CAAAGGGCCTGGGAGTTGAAGAAGGGTG TGGAGTAAGATGCTCATTCAAC[T/G]TGTTCTCAGAGTTAGAAAAAGGCCC AGGTAACCCAGGCCCCACAGA[G/A]AGGTGGTTGCATGCAAGTGTCTA TTTATTATTTTCTGGGTAAGAGAAGCAGCAC[T/C]GCTTTTATGTAACTTTTCCACTG CATGGTTCAATCTTGGTAGGTTGT[G/A]TGTATCTAGGAATTTATTCATTTATTCTAG TTTT TTTTTCATCTCTGATTATATTTATTTGG[A/G]TCTTCTCTCTTTTCTTCTTA GGCGTGAGCCACCACGCCTGGC[A/C]TGGATTTTTAAAAATTATTTCAATCTCCTTG TT TAGTAGGTTTCCTGGGGGATGAG[T/G]TCAAGGAATAAGAGGGGTCACTGACA CTCCGCCATCCACACTCCCAGGGTCTG[T/C]GGTGAAATCGAATTTATTCTCATTCT C AGGAGCTATTTTAATTTTCATGCAATGT[C/T]TCTGTAGGGTTCAGTTGGCGTTGT 10
11 11 12CTAF322 AGTATCTACCGTGTCTTAATAT[T/C]GTATCCAATTGGAATTTGATATAAAACT 13CTAF343 TGGGAGTATTTTACTGTGCTAAAA[T/C]ACATTTAGCATGGGCTGTTATATCTTATGA C 14CTAF371_1 CAATACTAACACAGATAGG[T/G]GTATCGTAGCAGAGTCCTGATTGTGCGCC 15rs GATAAAAAGGAACTGTTTAAA[C/T]TGATAGTAAAGAAAAGCCTTAAATTTTTGG 16CTAF422 GTTCTATGGTTAAGAATCCGTAAGCATCA[G/A]GTCCTTTATTTTGCATCTCTGGGG 17CTAF434_2 TCATTTAATTCATTACCCATATACTAC[G/A]TATGGGCATATAAATACCTGCTTT 18CTAF439-1 CAGGACCAAACAAAACTGAA[T/C]AGGAACTCAAGTCACTCAAGTAACTT 19CTAF439-2 CTTTGCTTCAAGACTCTCTTCT[C/G]TTTTATAGATGTAATTCCACTCACCCTCA 20CTAF450_1 GTTTAACCTTTTAGGAGGGTTTTGTTG[T/C]TGCCTTATAGAAGCTAACATAAG 21CTAF450_2 GCCAAAGAAAAGAAATGCAAAG[A/C]GGGCAGACAAAGAAAAAAAA 22CTAF450_3 GAAAAAAAAAAAAAACAC[G/A]TGAATGTAAAGCTATGCCACTTTTT 23CTAF450_4 AATGTAAAGCTATGCCACTTTTTT[T/A]CCCCTCTCATAGGTAATTCTGGAA 24MH30 AATAAAAACAGAATAAAACAAT[G/C]AGAAAATTTTCACCTTTATTTAATTAGCAGA 25MH26 CCCACTGACTCTGTCTCAAGAT[G/T]GTAATTAGTAACTGACAATGATTACGC 26MH23 TATCTAAGACTTGGATTCAGTG[C/A]ATTTCACATCGCTGAGCTCTCACAT 27MH20 TAGTCTGCCTGACATGCTT[G/A]GTGTATCTTGTATGATTTCTGTAAAGTTAGAA 28MH18 AAAATTGTTGAGAGTTGAAAGTAAGAGA[T/C]CCTTCCTAATAGTGCCTTCTTATCTC T 29MH17 CAATTTTATTTGCTAAATTTAGTAC[T/C]AGAGTGACATTATCTGTACATT 30MH15 TGCATACAGAAACTGAGCTCTATGC[G/T]TGTGCCAGACAAAAACCAAAGAGCTT 31MH14 CTGGCTGGTGATGGTTCAGTGT[G/A]ATGTGGGATCAGAATTGCAGATTAT 32MH13_2 ACCCGAAAAGGAAATCTCATAC[T/C]GCCAAGCAGTCATTCCCCATTTTCC 33MH13_1 CCTTCATTCCTTTTTATGGCT[G/A]AATAATATTCCGTGGTGTAGATAG 34MH3 AATTAAGGAATCTGGTAAGA[T/C]GTCTTTGAAAGTTCTAATTACAAC 35MH2 ATTTCCTTCTTTATAAAATGGGAAAAATG[G/A]TAACTCTTGTCTTGTAGGGTTGTTA 36MH1 AGTAGTGGCAACAGAGACCC[C/T]ACCGTTTGCAAATCATAACATATTTACT 37CT50 GCTCCTCTACATAATACTTCAA[T/C]TCCAGCATTGATCTCACTCTATCATGATCA 38CT51 CTCTATCATGATCATGGGTTTAGCTG[T/C]CTGTCCCTGCCACTGCTGTGTGT 39CT52 CAGGAACATTTGTTTTTCACTTTTT[A/G]AAAAACCTCTGTTGCCCAGTCT 40CT53 GTTGGCGCTTGAGCTGGGGCTTGAAG[G/A]TTTCTATAATGTGTAGCAGTGTAT 41CT41 AAAACCAGAGGCAGCTTCTTTTC[C/T]GCCTATTTTCAGTTTATTTCTTGTGAT 42 CT44 (- 319C>T) TCCACTTAGTTATCCAGATCCT[C/T]AAAGTGAACATGAAGCTTCAGT 43+49G>A GGCACAAGGCTCAGCTGAACCTGGCT[G/A]CCAGGACCTGGCCCTGCACT 44CT43 TATCTCTCTCTAGACCTTCTTGG[C/T]TAAGAAACCATGTAGTTTGTATGA 45CT55 GCAGCCACTATTTTTGAGTTGATGCAAG[T/C]CTCTCTGTATGGAGAGCTGGTCTCC 46CT57 GACAGTCCCTCTCAGACA[C/A]CTCTGCCTAAGGCCAGCTTTGCCATTGCA 47CT59 GTTTATAATTCTGTATGCTGTGA[A/G]CATTCATTTTTAACCAGCTAGGGACC 48CT60 TTTTGATTTCTTCACCACTATTTGGGATATAAC[G/A]TGGGTTAACACAGACATA 49CT61 TGGGTTAACACAGACATA[G/A]CAGTCCTTTATAAATCAATTGGCATGCTGT 50JO37_3 TGGTAGCCATGAAGAAAAACACCAATC[G/A]GGAGCCTCAGTGGATA 51JO37_2 GTATATCATTTCCACTCCTCTAAAC[G/A]TCTTTAGAGAGATTACTCTTTTTCATAGTT 52JO37_1 GGGAGTGCATCTGGGCACCA[A/C]ATGAAAGCCTCTTCTAATTCCCATGTCA 53JO36 GTCAGATTTGCTGACACTTTAAGCTC[A/G]TGGATTTCTCCTCTTTTGTTTCATAG 54JO35 TAGGACATCCAGGACCGTTTT[T/C]CATACAGAACCCATCTGTG 55JO34 CTACATGCTGGGATAGGGGCTCAT[A/G]GTAAGTTTGCCAGATTCAACCAA 56JO31 AACAGTCTGTCAGCAAAGCC[G/T]GCAGTACACTGAGAAAGCTCCTATT 57JO30 CGGACCTCTTGAGGTCAGGAGTTC[G/A]AGACCAGCCTGGCCAACATGGTGA 58JO27_2 CAGGGCCTGAAGATGCCCC[C/G]AAATGGCAAATTAAGTGAACTGG 59JO27_1 CCAGAAGTTGAAGTGTAGGAA[T/C]ATCTGGGGTCAAAGCAAAAAAAGACTTT 60JO26_2 GTGTGTGGTGTGTGTGTGGGCAC[C/T]AAGCAGGGGTGAAAGCTGATTATGGAGG 61JO26_1 GGTGAAAGCTGATTATGGAGGGG[T/C]GGTGTCAGGAAACATCACAAAGG 62JO23 AATGAGAGAGTGCTTACCAT[A/G]TTTGTCTACCCAAACCTTTAAAGA 63JO22 AATGTTGTACCCTTCAGCACA[T/C]TGGCATGAGCTCAGGTGGCAGGAGAACAA
12 12 64JO18 CTTTTACCTTTTCTTTATTTTCCCGTCAG[T/C]AAATATTTGTTAGGCAAATAAGAGCC 65JO13 TGGCAAATCTGGTCACCACAA[T/C]ACTCTTTAAAAAACACGCTCATGTTATT 66JO10 CTCCCAGAGTTTGCCCTGTCCAAG[G/T]AATTCAGGGGAAAACGATGGTGGGCTC 67JO9 CATGTAATTAGCCTACTGGCTCTA[C/G]AATAGTCATGATATATCCATTAAAA 68JO8_2 AGAAAGACCCAAACCCACTTTTATACCAAACCCAC[T/C]CTTGTGATAACAAA 69JO8_1 TTGTGATAACAAAC[T/C]CACTCCTGTTCTAATGACAGAGCCCTCATGACCT 70JO6_2 CTAACAATCTGAAATGTGAAA[T/C]TATCTCRCTTTCACACAAGAAG 71JO6_1 CAATCTGAAATGTGAAAYTATCTC[A/G]CTTTCACACAAGAAGAGATGTAGAA 72JO3 AATATTAACCCTCAACAAC[C/A]GTAATGGATATAAACGCATGTGCTCATCA 73CTBC358 GAAGGTGGTGCTCAGAGCAG[G/T]CTGCTTCCCTGCCAGGGTTATAGTCC 74CTBC313 CTCAGTTTCAGAAATGTCCTCT[C/T]ATAGAAGCCCCTAGGAAAACAAGCCT 75CTBC305 TCTGGTTTGCGTAATTTATGC[C/T]TTTGAAAGGCTGATATTATTGCATT 76CTBC217_2 ATTTCTTTCTGGATATTGAAAA[A/C]CAATAGGGGTAGTGYCTTCCTTC 77CTBC217_1 GAAAAMCAATAGGGGTAGTG[C/T]CTTCCTTCTAACCTCAGGGGATCCCTTCAG 78CTBC190 GCAGCTATGTTTCAGATGTAATT[T/C]TGTTTCCACCAATCAGATGTGCCGAT 79CTBC182_2 ACACTTCAGTGCATCTCTAATTT[G/T]GGAAATGATCAATGGTCAACAGAG 80CTBC182_1 TAGTAGTTTTTACAATTGTATT[C/T]ATTAGGCACAGGAAAATGTAAACTCAG 81CTBC165_3 AAGGGCTGGAGAAGCAGATT[A/C]TTGGGCTTAGCTTCTAGAAATA 82CTBC165_2 AGTTCCAAAAGCAAGTGAGGTATTGGCA[C/T]ATCTGATTGGTGGAAACAAAATTA 83CTBC165_1 ACAAAATTACATCTGAATTTTGTTT[G/T]CTGAATTGCTACATGGGCATAGG 84CTBC106 TTCTTGACCAAGATTTTTGAGAAGG[C/T]GAGGCATGGGAAGGATCATTG 85CTBC099 GGGGAAGCTTCAGCAGCTAGG[G/A]GGATCCCTGCAGGATCAGCCT 86CTBC078 CATTTGGACTGGGAATTCTT[T/C]CTGTGGTAGAAAATAAAATG 87CTBC073 TTAAGTGCAGTTAAGAGGA[T/C]TGAGGAATGTAGAATGAGCACAAG 88CTBC053 GAAGTCTGTGAAGATGACTGACC[T/C]TGGAGCCCCTCTTAGCCTCTCCAC 89CIIC065 AAAGCAACAGTGGGGCAACATATTT[T/C]ACTGTCAGACAAATAAAGCCC 90IC082R ATGAAAATTTGTCATATTAAAAAG[G/A]AAAATAAAACAAAAAGAA 91CTIC098 TGCCTCCAATATGAAAGCTAC[C/T]AAAGGACAGAGGCCATACCAGTTT 92CTIC114_1 GACCAACAGGGAGACAATT[C/T]CTTCCCCCSAAGACATACCTACTAAT 93CTIC114_2 CCAACAGGGAGACAATTYCTTCCCCC[C/G]AAGACATACCTACTAATTAAAC 94CTIC114_3 ATTTTTAAAAAATTATTATGC[A/G]TGGGCTGCGAATTTGAATCCAAGGTTCCTGA 95CTIC142_1 TGTAGGGAACTGGCACATGGAGAGCATTTAGA[T/G]AATTTATGCTGAATTTTTGTTA C 96CTIC142_2 GATGTGACCCTATAAT[A/C]TGGAACTCTGGCACCCAGGCATG 97CTIC142_3 AAACAAACACATTTACAA[G/A]AAAAATGTTTTAAAGATGCCAGGGGTACT 98CTIC148 AGTTAAAATGGTTTACTTTGTTCA[A/T]GTTAGTGGTAGGAAACATTGCCC 99CTIC154_1 TCTTTAATGGGCCAGCATTCT[C/T]ATGGGGTAGAGCAGAATATTC 100CTIC154_2 GAATCACAGTGGTCTACCTGC[A/G]TTCATAATTCCAGGATCTGTGAA 101CTIC154_3 TGGCTAGAAAGATTCTTAAATAT[G/A]TGGAATATGATTATTCTTAGCTGGAATATTT 102CTIC159 TGTGTGTGGGGTGGGGTATGGGGAGGA[G/C]AACCTTCATGGTGGCCCACCTGG 103JC034sR CATTCAAAGAAATGATGGTCC[G/A]TCAGAAATACATTTTAACTGAG 104JC058sR ATGGTATATGTTAAATGCT[G/A]TGGTGTCTGAATTTATTTCCTAGCAACA 105JC068sFa ATCCCTAAATATTCATTCTCCT[C/G]TCTTATCTCCCATCTTCCTCTA 106JC068sRb CTGCAACTCCAGCAGGTAAAATCTG[C/G]TTACCTAACCACCAATTGTGGTATGG 107JC473sR_4 GTTTTCCTAGAATTGATTC[A/C]ATTAACAACTTACTGGGGTTTTAAAAG 108JC569sF GTAACAAGGAAAACTCTACAGGGTGG[G/A]ATTGAGATATTGGATCTTGATATTA
13 DNA preparation For cell-line samples, DNA was extracted from Epstein-Barr virus (EBV)- transformed peripheral blood lymphocytes. One hundred ml of confluent EBV cells were pelleted at 1000 rpm for 5 min. The supernatant was discarded and cells were re-suspended in 3.5 ml of 6 M guanidine hydrochloride (Sigma, Dorset, UK), 250 µl of 7.5 M ammonium acetate (VWR international Ltd, Lutterworth, UK), 50 µl of Proteinase K (VWR international Ltd, Lutterworth, UK) and 250 µl of 20% sodium N-laurylsarcosinate solution (Fluka, Dorset, UK). The solution was incubated overnight at 37 C. The tubes were cooled to RT and then 2 ml of chloroform added. The samples were then mixed and spun at 2500 rpm for 3 min. The upper layer was removed and added to 10 ml of absolute ethanol. The tubes were chilled to -20 C overnight. The solution was gently inverted and the precipitated DNA pelleted at 3000 rpm for 15 min in an Allegra 6R micro-centrifuge (Beckman, High Wycombe,UK). Pellets were washed with 70% ethanol and re-suspended in 300 l TE (10 mm Tris, 0.1 mm EDTA; ph 7.5). DNA was quantified using PicoGreen dsdna quantitation reagent (Molecular Probes Europe B.V., Leiden, The Netherlands) and diluted to 4 ng/ul in TE (ph 7.5) before use. For blood samples, whole blood was lysed using 30 ml of lysis buffer (0.3 M Sucrose, 1.0 M Tris-HCl, 4.9 M MgCl 2, 1% Triton-X (Sigma, Dorset, UK)). DNA was extracted from the resultant white blood cell pellet using the method described above for cell-line samples. Genotyping PCR PCR conditions were optimised by varying MgCl 2 concentrations between 1 and 5 mm and annealing temperature between 50 C and 65 C. PCRs were performed in 384 well polypropylene microtitre plates (ABgene, Epsom, Surrey, UK) in 6 µl final 13
14 reaction volume. 3 µl of 4 ng/µl stock of genomic DNA was dispensed into each well. 3 µl of PCR reaction mix containing 0.4 mm dntp, 2-10 mm MgCl 2, 3.8 ng/µl forward and reverse primer, and 0.25 units of TaqGold (Applied Biosystems, Foster City, CA) was added and the plate was covered with Adhesive Sealing Sheet (ABgene, Epsom, Surrey, UK). Reactions were incubated at 95 o C for 15 min and then cycled for 35 cycles of 95 o C for 30 s, o C for 30 s, 72 o C for 30 s, and finally incubated for 15 min at 72 o C on PCT225 thermocyclers (MJ Research, Watertown, MA) using heated lids. All pipetting steps for PCR preparation were performed with on a Beckman Multimek 96 robot (Beckman, High Wycombe, UK), dedicated to pre-pcr work. Genotyping methods Apart from early RFLPs 5 all the genotyping on this project was performed using the Invader genotyping method described previously 6. Probe sets for each SNP were designed and synthesised by Third Wave Technologies, Inc (Madison, WI). For a uniplex dry-down plate Invader assay, assays were prepared for each allele separately. In a 384-well format, PCR products were diluted 1:4 in distilled water. Aliquots of 3 µl were dispensed into two pre-prepared 384-well format Invader Assay FRET detection plates (Third Wave Technologies, Inc. Madison, WI). Six microlitres of probe mix containing 1.3 µm allele specific probe, 0.13 µm Invader TM probe, and 7.5 mm MgCl 2 were added. All pipetting steps for this preparation were performed with a Beckman Multimek 96 robot. Plates were covered with Adhesive Sealing Sheet (ABgene, Epsom, Surrey, UK). Reactions were incubated at 95 o C for 5 min, and 65 o C for min on MJ PCT225 thermocyclers. Fluorescence was measured directly at the end of incubation using a Cytofluor 4400 fluorescence 14
15 microtiter plate reader (Perkin Elmer Applied Biosystems, Foster City, CA), excitation 485/20, emission 530/25, and gain 50. For a biplex dry-down plate Invader assay, assays for both SNP alleles were performed at the same time. Final reaction concentrations of the allele-specific probes were 0.7 µm and the Invader TM probe was 0.07 µm. The probes were labelled either FAM or RED. Fluorescence was measured directly at the end of incubation using a Cytofluor 4400, excitation 485/20, emission 530/25 for FAM and excitation 560/20, emission 620/40 for RED. All pipetting steps were performed with a Beckman Multimek 96 robot. The (AT) n -3 UTR microsatellite marker was genotyped using fluorescentlylabelled primers as described elsewhere 7. Genotype concordance Genotyping concordance was assessed by double-typing a small number of key SNPs using either a second batch of biplex Invader kit, or the Taqman MGB chemistry (Applied Biosystems Inc, Foster City, CA) 8-9. MH30, CT60 and JO31 were genotyped using both Taqman and Invader in either UK T1D families, GD cases or controls by two scientists independently. The allelic differences were 0/632(0%) for MH30, 1/688 (0.037%) for CT60, 1/1,898 (0.053%) for JO31, in total, 2/5,218 (0.038%). CTAF371_1 was genotyped using Taqman and biplex Invader by the same scientist. No allelic difference was observed in 1,990 alleles. No typing difference was observed in 768 alleles for CTIC154_1 comparing two different batches of biplex Invader kits. Genotype checks 15
16 All the genotyping data was stored in our local MySQL database ( This allows easy double scoring, error checking and data retrieval. Data manipulation errors are reduced to a minimum by employing automated approaches. All Invader and Taqman data was scored twice independently. Case-control datasets In the case-control datasets, genotyping data was checked for Hardy-Weinberg equilibrium (HWE) separately for the cases and controls. Markers and datasets showing deviation from HWE (0.01< P <0.05) are presented in Table A4. Datasets, which showed deviation from HWE (P< 0.01) were treated as genotyping errors, and were retyped. No data set used in analysis showed deviation from HWE (P < 0.01). Of the 216 datasets (108 SNPs) typed in the Graves cases and controls, 18 datasets are out of HWE (0.01< P <0.05). No datasets were out of HWE in the large Graves case-control dataset or in the AIH cases. Since regression analyses are sensitive to the number of typed samples, we retyped SNPs if there were > 10% genotyping failures in either the cases or controls. The percentages of genotype failures were as a result less than 10% in each case and control dataset used, except for rs v cases (15.9%). D (Ref) 10 and W n (Ref) 11 were used as the measure of pairwise LD in the LD blocks, measured both in a moving window across the region and against the doubletyped key frame markers, MH30 and CT60. If D or W n differed substantially between cases and controls between a pair of markers (and D > 0.8), we retyped the marker. 16
17 Table A4: Markers and datasets showing deviation from Hardy-Weinberg equilibrium (a) Graves cases and controls Marker Dataset P value MH13_1 cases MH2 cases MH1 cases IC082R cases CTIC098 cases CTIC159 cases JC068sFa cases rs controls rs controls MH23 controls CT43 controls CT61 controls CTBC106 controls CTBC099 controls CTBC073 controls CTBC053 controls JC058sR controls JC473sR_4 controls (b) T1D families CTAF343 USA Family collections All typing data on family collections, including the microsatellite marker (AT) n 3 UTR, were screened for Mendelian inconsistencies using PedCheck 12. Families that consistently misinherit in > 3% of SNPs were marked in the database, and automatically excluded from analysis. The familial dataset was screened again for Mendelian inconsistencies in STATA ( The working hypothesis is that these give a measure of genotyping error, given we have already excluded "real" misinheritances. These apparent misinheritances were eliminated by re-typing or excluding a family from the analysis for that marker. Mendelian inconsistencies in the 3,671 T1D family dataset are tabulated in Table A5. There were 17
18 231 inconsistencies out of 41,406 informative transmissions (0.56%). Some families identified by STATA have been recently excluded as newly-identified consistently mis-inheriting families in our lab. The number of apparent offspring inconsistencies arising from genotyping errors is, therefore, less than 0.56%. D and W n values were calculated with two double-typed frame markers (MH30, CT60) and were used for LD pattern checking. If the datasets showed largely different D or W n values within each population, we re-typed the marker. The familial genotyping data was also screened for HWE using parents only by population, grouped as UK (Warren I, Yorkshire, Belfast, South West and Bristol), Norway, Romania, USA and Finland. Only one of 41 datasets showed deviation from HWE in Graves and T1D families (not shown). Table A5: Apparent misinheritances in 3,671 T1D families Affected sibling Unaffected SNP name Inconsistencies Informative % Inconsistencies Informative % Total % CTAF % rs % MH % +49G>A % CT % JO % JO % JO27_ % CTIC % Total % CTLA-4 gene expression Primers and probes to detect human CTLA-4 and sctla-4 mrnas were designed using Primer Express software (Applied Biosystems, Foster City, CA), β2- microglobulin by use of Primer 3 program (Table A6). The mouse primer and probe sets are shown in Table A7. All the synthesised probes were labelled FAM and 18
19 TAMRA at the 5 and 3, respectively. All the sequences and the annealing temperatures used for the 3 UTR mapping are tabulated in Table A8. Figure A4 shows our transcription analysis results of human the CTLA-4 gene. Table A6: Primers and probes for human Taqman assay Target Gene Forward (5-3 ) Reverse (5-3 ) Probe (5-3 ) flctla-4 ACCCAGATTTATGTAATTG ATCCAGAA CCGAACTAACTGCTGCAAGG A CGTGCCCAGATTCTGACTTCCT CCTCT sctla-4 CATCTGCAAGGTGGAGCT CAT GGCTTCTTTTCTTTAGCAATT ACATAAATC ACCGCCATACTACCTGGGCAT AGGCA β2 microglobulin TGCTCGCGCTACTCTCTCT TCCATTCTCTGCTGGATGAC CTGGAGGCTATCCAGCGTACT CCAA Table A7: Primers and probes for mouse Taqman assay Target Gene Forward (5-3 ) Reverse (5-3 ) Probe (5-3 ) flctla-4 ACTCATGTACCCACCGC CATA GGGCATGGTTCTGGATCAAT CATGGGCAACGGGACGCAGAT TTAT sctla-4 lictla-4 ACCCACCGCCATACTTT GTG AGGACTTCTTTTCTTTAGCA ATGACAT GCCTTTTGTAGCCCTGCT TCAGAATCCGGGCATGGTT CA AATCTGCGTCCCGTTGCCCATG C TTCTTTTCATCCCAGTCTTCTCT GAAGATCCA β2 microglobulin CCTGCAGAGTTAAGCAT GCCA TGATCACATGTCTCGATCCC A TATGGCCGAGCCCAAGACCGT CT 19
20 Table A8: Primers for the 3 UTR mapping Forward (5-3 ) Reverse (5-3 ) PCR condition (annealing) GAGCTGGGATGTTTCTGTCA CTTAATTCCTTGACCCACATCA 55 C. ATGAGTCAGCTTTGCACCAGC CATTAC GAGGTGAAGAACCTGTGTTAAACAG CATG TD 70 C GAACCACTTGTCACAGGCAAG ACTG GTCAGCAAATCTGACGGTGGAGGC TD 70 C TGTGATTATCACCACTGTTGCT GCT CTGTTAGTTGCTATAGTAACTTGACT G 55 C CTGCTTCCACCTTGTCATGTTA GCCG GTAGAGACAGGGTTTCACCATGTTGG C TD 70 C CTGCTTCCACCTTGTCATGTTA GCCG TCTCCTGCCTCAGCCACCCGAGTAGC G TD 70 C CATAGTCTCTACTCTCCTGTGA GC ATGGAGATGATGGCAGAACCTC 55 C. 20
21 Figure A4: Transcription of the human CTLA-4 gene. Seven different sets of primers, indicated by open boxes, were designed on the basis of the genomic sequence downstream the open reading frame of CTLA4. For each primer set both +RT (reverse transcriptase)(+) and RT(-) PCR products are displayed. RT-PCR mapping placed the end of the 3 UTR 6.57 kb 3 the last codon of CTLA-4 and CT60, JO31 and JO30 inside the 3 UTR. Northern Blot of poly(a)+ mrna derived from unstimulated and 24 h stimulated PBMC show the presence of multiple transcripts of CTLA-4 due to the existence of different poly(a) signal (A-G) in the 3 UTR. JO31 and JO30 are only present in the long 6.7 kb transcript. All five of the kb transcripts include CT60. Primer sequences are given in Table A8. With respect to the pattern of splicing since the disease-associated 6.1 kb region is only 279 bp 3 of the major polyadenylation poly(a) mrna termination site of CTLA4, we propose that its allelic variation might affect termination of the transcript. Sequences 3 of the poly(a) site bind the protein complex that mediates transcription termination The termination 21
22 protein complex in turn binds to the proteins mediating splicing of the final exon in the highly orchestrated production of a mature transcript 15. The pre-mrna complex destined to form mature sctla-4 mrna may be more stable when the protective allele contributes to the secondary structure or, alternatively, the splicing needed to produce the sctla-4 mrna from the protective allele may be more efficient. References 1. Osoegawa, K. et al. A bacterial artificial chromosome library for sequencing the complete human genome. Genome Res 11, (2001). 2. Osoegawa, K. et al. Bacterial artificial chromosome libraries for mouse sequencing and functional analysis. Genome Res 10, (2000). 3. Durbin, R. & Thierry-Mieg, J. AC.elegans database. Documentation, code and data available from anonymous FTP servers at lirmm.limrmm.fr, cele.mrclbm.cam.ac.uk and ncbi.nlm.nih.gov. (1991). 4. Schwartz, S. et al. PipMaker--a web server for aligning two genomic DNA sequences. Genome Res 10, (2000). 5. Johnson, G. C. et al. Haplotype tagging for the identification of common disease genes. Nat Genet 29, (2001). 6. Mein, C. A. et al. Evaluation of single nucleotide polymorphism typing with Invader on PCR amplicons and its automation. Genome Res 10, (2000). 7. Nistico, L. et al. The CTLA-4 gene region of chromosome 2q33 is linked to, and associated with, type 1 diabetes. Hum Mol Gen 5, (1996). 8. User Bulletin: Primer Express Version 1.5 and TaqMan MGB Probes for Allelic Discrimination ( 22
23 9. Ranade, K. et al. High-Throughput Genotyping with Single Nucleotide Polymorphisms. Genome Res.11, (2001) 10. Devlin, B. & Risch, N. A comparison of linkage disequilibrium measures for fine-scale mapping. Genomics 29, (1995). 11. Klitz, W., Stephens, J. C., Grote, M. & Carrington, M. Discordant patterns of linkage disequilibrium of the peptide- transporter loci within the HLA class II region. Am J Hum Genet 57, (1995). 12. O Connell, J. R. & Weeks, D. E. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63, (1998). 13. Proudfoot, N. J., Furger, A. & Dye, M. J. Integrating mrna processing with transcription. Cell 108, (2002). 14. Dye, M. J. & Proudfoot, N. J. Multiple transcript cleavage precedes polymerase release in termination by RNA polymerase II. Cell 105, (2001). 15. Cartegni, L., Chew, S. L. & Krainer, A. R. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet 3, (2002). 23
MALE ADULT FIBROBLAST LINE (82-6hTERT)
 Double-stranded methylation patterns of a 104-bp L1 promoter in DNAs from male and female fibroblasts, male leukocytes and female lymphoblastoid cells using hairpin-bisulfite PCR. Fifteen L1 sequences
Double-stranded methylation patterns of a 104-bp L1 promoter in DNAs from male and female fibroblasts, male leukocytes and female lymphoblastoid cells using hairpin-bisulfite PCR. Fifteen L1 sequences
tgg agg Supplementary Figure S1.
 ttaggatattcggtgaggtgatatgtctctgtttggaaatgtctccgccattaactcaag tggaaagtgtatagtaatgaatctttcaagcacacagatcacttcaaaagactgtttcaa catcacctcaggacaaaaagatgtactctcatttggatgctgtgatgccatgggtcacag attgcaattcccaagtgcccgttcttttacaccaaaatcaaagaagaatatctccccttt
ttaggatattcggtgaggtgatatgtctctgtttggaaatgtctccgccattaactcaag tggaaagtgtatagtaatgaatctttcaagcacacagatcacttcaaaagactgtttcaa catcacctcaggacaaaaagatgtactctcatttggatgctgtgatgccatgggtcacag attgcaattcccaagtgcccgttcttttacaccaaaatcaaagaagaatatctccccttt
FETAL FIBROBLASTS, PASSAGE 10
 Double-stranded methylation patterns of a 104-bp L1 promoter in DNAs from fetal fibroblast passages 10, 14, 17, and 22 using barcoded hairpinbisulfite PCR. Fifteen L1 sequences were analyzed for passages
Double-stranded methylation patterns of a 104-bp L1 promoter in DNAs from fetal fibroblast passages 10, 14, 17, and 22 using barcoded hairpinbisulfite PCR. Fifteen L1 sequences were analyzed for passages
Experimental Identification and Computational Characterization of a Novel. Extracellular Metalloproteinase Produced by Clostridium sordellii
 Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 207 Supplementary Information Experimental Identification and Computational Characterization of
Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 207 Supplementary Information Experimental Identification and Computational Characterization of
Functional Genomics & Proteomics
 Functional Genomics & Proteomics Genome Sequences TCACAATTTAGACATCTAGTCTTCCACTTAAGCATATTTAGATTGTTTCCAGTTTTCAGCTTTTATGACTAAATCTTCTAAAATTGTTTTTCCCTAAATGTATATTTTAATTTGTCTCAGGAGTAGAATTTCTGAGTCATAAAGCGGT CATATGTATAAATTTTAGGTGCCTCATAGCTCTTCAAATAGTCATCCCATTTTATACATCCAGGCAATATATGAGAGTTCTTGGTGCTCCACATCTTAGCTAGGATTTGATGTCAACCAGTCTCTTTAATTTAGATATTCTAGTACAT
Functional Genomics & Proteomics Genome Sequences TCACAATTTAGACATCTAGTCTTCCACTTAAGCATATTTAGATTGTTTCCAGTTTTCAGCTTTTATGACTAAATCTTCTAAAATTGTTTTTCCCTAAATGTATATTTTAATTTGTCTCAGGAGTAGAATTTCTGAGTCATAAAGCGGT CATATGTATAAATTTTAGGTGCCTCATAGCTCTTCAAATAGTCATCCCATTTTATACATCCAGGCAATATATGAGAGTTCTTGGTGCTCCACATCTTAGCTAGGATTTGATGTCAACCAGTCTCTTTAATTTAGATATTCTAGTACAT
1. SIT. The handler and dog stop with the dog sitting at heel. When the dog is sitting, the handler cues the dog to heel forward.
 START START SIT 1. SIT. The handler and dog stop with the dog sitting at heel. When the dog is sitting, the handler cues the dog to heel forward. This is a static exercise. SIT STAND 2. SIT STAND. The
START START SIT 1. SIT. The handler and dog stop with the dog sitting at heel. When the dog is sitting, the handler cues the dog to heel forward. This is a static exercise. SIT STAND 2. SIT STAND. The
Methods S1. Sequences relevant to the constructed strains, Related to Figures 1-6.
 Methods S1. Sequences relevant to the constructed strains, Related to Figures 1-6. A. Promoter Sequences Gal4 binding sites are highlighted in the color referenced in Figure 1A when possible. Site 1: red,
Methods S1. Sequences relevant to the constructed strains, Related to Figures 1-6. A. Promoter Sequences Gal4 binding sites are highlighted in the color referenced in Figure 1A when possible. Site 1: red,
16. Allocation Models
 16. Allocation Models Juha Saloheimo 17.1.27 S steemianalsin Optimointiopin seminaari - Sks 27 Content Introduction Overall Efficienc with common prices and costs Cost Efficienc S steemianalsin Revenue
16. Allocation Models Juha Saloheimo 17.1.27 S steemianalsin Optimointiopin seminaari - Sks 27 Content Introduction Overall Efficienc with common prices and costs Cost Efficienc S steemianalsin Revenue
Supporting Information for
 Supporting Information for Analysis of Sogatella furcifera proteome that interact with P10 protein of Southern rice black-streaked dwarf virus Win Than*, Faliang Qin*, Wenwen Liu, Xifeng Wang ** State
Supporting Information for Analysis of Sogatella furcifera proteome that interact with P10 protein of Southern rice black-streaked dwarf virus Win Than*, Faliang Qin*, Wenwen Liu, Xifeng Wang ** State
Capacity Utilization
 Capacity Utilization Tim Schöneberg 28th November Agenda Introduction Fixed and variable input ressources Technical capacity utilization Price based capacity utilization measure Long run and short run
Capacity Utilization Tim Schöneberg 28th November Agenda Introduction Fixed and variable input ressources Technical capacity utilization Price based capacity utilization measure Long run and short run
Supporting information
 Supporting information Figure S1. Carotenoid biosynthesis pathway in papaya fruit which is adopted from Blas et al. (2010) (reference 4) and Nisar et al. (2015) (reference 5). Carotenoids are synthesized
Supporting information Figure S1. Carotenoid biosynthesis pathway in papaya fruit which is adopted from Blas et al. (2010) (reference 4) and Nisar et al. (2015) (reference 5). Carotenoids are synthesized
State of the Union... Functional Genomics Research Stream. Molecular Biology. Genomics. Computational Biology
 Functional Genomics Research Stream State of the Union... Research Meeting: February 16, 2010 Functional Genomics & Research Report III Concepts Genomics Molecular Biology Computational Biology Genome
Functional Genomics Research Stream State of the Union... Research Meeting: February 16, 2010 Functional Genomics & Research Report III Concepts Genomics Molecular Biology Computational Biology Genome
Basset: Learning the regulatory code of the accessible genome with deep convolutional neural networks. David R. Kelley
 Basset: Learning the regulatory code of the accessible genome with deep convolutional neural networks David R. Kelley DNA codes for complex life. How? Kundaje et al. Integrative analysis of 111 reference
Basset: Learning the regulatory code of the accessible genome with deep convolutional neural networks David R. Kelley DNA codes for complex life. How? Kundaje et al. Integrative analysis of 111 reference
Plasmid Name: pmm290. Aliases: none known. Length: bp. Constructed by: Mike Moser/Cristina Swanson. Last updated: 17 August 2009
 Plasmid Name: pmm290 Aliases: none known Length: 11707 bp Constructed by: Mike Moser/Cristina Swanson Last updated: 17 August 2009 Description and application: This is a mammalian expression vector for
Plasmid Name: pmm290 Aliases: none known Length: 11707 bp Constructed by: Mike Moser/Cristina Swanson Last updated: 17 August 2009 Description and application: This is a mammalian expression vector for
Supplementary information: Biocatalysis on the surface of Escherichia coli: melanin pigmentation of the cell. exterior
 Supplementary information: Biocatalysis on the surface of Escherichia coli: melanin pigmentation of the cell exterior Martin Gustavsson, David Hörnström, Susanna Lundh, Jaroslav Belotserkovsky, Gen Larsson
Supplementary information: Biocatalysis on the surface of Escherichia coli: melanin pigmentation of the cell exterior Martin Gustavsson, David Hörnström, Susanna Lundh, Jaroslav Belotserkovsky, Gen Larsson
Returns to Scale II. S ysteemianalyysin. Laboratorio. Esitelmä 8 Timo Salminen. Teknillinen korkeakoulu
 Returns to Scale II Contents Most Productive Scale Size Further Considerations Relaxation of the Convexity Condition Useful Reminder Theorem 5.5 A DMU found to be efficient with a CCR model will also be
Returns to Scale II Contents Most Productive Scale Size Further Considerations Relaxation of the Convexity Condition Useful Reminder Theorem 5.5 A DMU found to be efficient with a CCR model will also be
Use of spatial data in the new production environment and in a data warehouse
 Use of spatial data in the new production environment and in a data warehouse Nordic Forum for Geostatistics 2007 Session 3, GI infrastructure and use of spatial database Statistics Finland, Population
Use of spatial data in the new production environment and in a data warehouse Nordic Forum for Geostatistics 2007 Session 3, GI infrastructure and use of spatial database Statistics Finland, Population
Efficiency change over time
 Efficiency change over time Heikki Tikanmäki Optimointiopin seminaari 14.11.2007 Contents Introduction (11.1) Window analysis (11.2) Example, application, analysis Malmquist index (11.3) Dealing with panel
Efficiency change over time Heikki Tikanmäki Optimointiopin seminaari 14.11.2007 Contents Introduction (11.1) Window analysis (11.2) Example, application, analysis Malmquist index (11.3) Dealing with panel
Eukaryotic Comparative Genomics
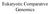 Eukaryotic Comparative Genomics Detecting Conserved Sequences Charles Darwin Motoo Kimura Evolution of Neutral DNA A A T C TA AT T G CT G T GA T T C A GA G T A G CA G T GA AT A GT C T T T GA T GT T G T
Eukaryotic Comparative Genomics Detecting Conserved Sequences Charles Darwin Motoo Kimura Evolution of Neutral DNA A A T C TA AT T G CT G T GA T T C A GA G T A G CA G T GA AT A GT C T T T GA T GT T G T
Metsälamminkankaan tuulivoimapuiston osayleiskaava
 VAALAN KUNTA TUULISAIMAA OY Metsälamminkankaan tuulivoimapuiston osayleiskaava Liite 3. Varjostusmallinnus FCG SUUNNITTELU JA TEKNIIKKA OY 12.5.2015 P25370 SHADOW - Main Result Assumptions for shadow calculations
VAALAN KUNTA TUULISAIMAA OY Metsälamminkankaan tuulivoimapuiston osayleiskaava Liite 3. Varjostusmallinnus FCG SUUNNITTELU JA TEKNIIKKA OY 12.5.2015 P25370 SHADOW - Main Result Assumptions for shadow calculations
Tynnyrivaara, OX2 Tuulivoimahanke. ( Layout 9 x N131 x HH145. Rakennukset Asuinrakennus Lomarakennus 9 x N131 x HH145 Varjostus 1 h/a 8 h/a 20 h/a
 , Tuulivoimahanke Layout 9 x N131 x HH145 Rakennukset Asuinrakennus Lomarakennus 9 x N131 x HH145 Varjostus 1 h/a 8 h/a 20 h/a 0 0,5 1 1,5 km 2 SHADOW - Main Result Assumptions for shadow calculations
, Tuulivoimahanke Layout 9 x N131 x HH145 Rakennukset Asuinrakennus Lomarakennus 9 x N131 x HH145 Varjostus 1 h/a 8 h/a 20 h/a 0 0,5 1 1,5 km 2 SHADOW - Main Result Assumptions for shadow calculations
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
( ( OX2 Perkkiö. Rakennuskanta. Varjostus. 9 x N131 x HH145
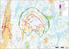 OX2 9 x N131 x HH145 Rakennuskanta Asuinrakennus Lomarakennus Liike- tai julkinen rakennus Teollinen rakennus Kirkko tai kirkollinen rak. Muu rakennus Allas Varjostus 1 h/a 8 h/a 20 h/a 0 0,5 1 1,5 2 km
OX2 9 x N131 x HH145 Rakennuskanta Asuinrakennus Lomarakennus Liike- tai julkinen rakennus Teollinen rakennus Kirkko tai kirkollinen rak. Muu rakennus Allas Varjostus 1 h/a 8 h/a 20 h/a 0 0,5 1 1,5 2 km
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.9.269
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.9.269
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Calculation: N117 x 9 x HH141 Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG
SHADOW - Main Result Calculation: N117 x 9 x HH141 Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG
Supporting Information for Electrophoresis DOI /elps Sunil Archak, Vemireddy Lakshminarayanareddy and Javaregowda Nagaraju
 ELECTROPHORESIS Supporting Information for Electrophoresis DOI 10.1002/elps.200600646 Sunil Archak, Vemireddy Lakshminarayanareddy and Javaregowda Nagaraju High-throughput multiplex microsatellite marker
ELECTROPHORESIS Supporting Information for Electrophoresis DOI 10.1002/elps.200600646 Sunil Archak, Vemireddy Lakshminarayanareddy and Javaregowda Nagaraju High-throughput multiplex microsatellite marker
Tork Paperipyyhe. etu. tuotteen ominaisuudet. kuvaus. Väri: Valkoinen Malli: Vetopyyhe
 etu Monikäyttöpaperi hoitaa useimmat pyyhintätehtävät Sopiva lasipintojen pyyhintään Sopii käsien kuivaamiseen Elintarvikekäyttöön hyväksytty Tork Easy Handling, pakkaus, jota on helppo kantaa mukana,
etu Monikäyttöpaperi hoitaa useimmat pyyhintätehtävät Sopiva lasipintojen pyyhintään Sopii käsien kuivaamiseen Elintarvikekäyttöön hyväksytty Tork Easy Handling, pakkaus, jota on helppo kantaa mukana,
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
Other approaches to restrict multipliers
 Other approaches to restrict multipliers Heikki Tikanmäki Optimointiopin seminaari 10.10.2007 Contents Short revision (6.2) Another Assurance Region Model (6.3) Cone-Ratio Method (6.4) An Application of
Other approaches to restrict multipliers Heikki Tikanmäki Optimointiopin seminaari 10.10.2007 Contents Short revision (6.2) Another Assurance Region Model (6.3) Cone-Ratio Method (6.4) An Application of
Mat Seminar on Optimization. Data Envelopment Analysis. Economies of Scope S ysteemianalyysin. Laboratorio. Teknillinen korkeakoulu
 Mat-2.4142 Seminar on Optimization Data Envelopment Analysis Economies of Scope 21.11.2007 Economies of Scope Introduced 1982 by Panzar and Willing Support decisions like: Should a firm... Produce a variety
Mat-2.4142 Seminar on Optimization Data Envelopment Analysis Economies of Scope 21.11.2007 Economies of Scope Introduced 1982 by Panzar and Willing Support decisions like: Should a firm... Produce a variety
The CCR Model and Production Correspondence
 The CCR Model and Production Correspondence Tim Schöneberg The 19th of September Agenda Introduction Definitions Production Possiblity Set CCR Model and the Dual Problem Input excesses and output shortfalls
The CCR Model and Production Correspondence Tim Schöneberg The 19th of September Agenda Introduction Definitions Production Possiblity Set CCR Model and the Dual Problem Input excesses and output shortfalls
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table 22.12.2014 11:33 / 1 Minimum
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table 22.12.2014 11:33 / 1 Minimum
TM ETRS-TM35FIN-ETRS89 WTG
 VE1 SHADOW - Main Result Calculation: 8 x Nordex N131 x HH145m Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please
VE1 SHADOW - Main Result Calculation: 8 x Nordex N131 x HH145m Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please
Alternative DEA Models
 Mat-2.4142 Alternative DEA Models 19.9.2007 Table of Contents Banker-Charnes-Cooper Model Additive Model Example Data Home assignment BCC Model (Banker-Charnes-Cooper) production frontiers spanned by convex
Mat-2.4142 Alternative DEA Models 19.9.2007 Table of Contents Banker-Charnes-Cooper Model Additive Model Example Data Home assignment BCC Model (Banker-Charnes-Cooper) production frontiers spanned by convex
Gap-filling methods for CH 4 data
 Gap-filling methods for CH 4 data Sigrid Dengel University of Helsinki Outline - Ecosystems known for CH 4 emissions; - Why is gap-filling of CH 4 data not as easy and straight forward as CO 2 ; - Gap-filling
Gap-filling methods for CH 4 data Sigrid Dengel University of Helsinki Outline - Ecosystems known for CH 4 emissions; - Why is gap-filling of CH 4 data not as easy and straight forward as CO 2 ; - Gap-filling
( ,5 1 1,5 2 km
 Tuulivoimala Rakennukset Asuinrakennus Liikerak. tai Julkinen rak. Lomarakennus Teollinen rakennus Kirkollinen rakennus Varjostus "real case" h/a 1 h/a 8 h/a 20 h/a 4 5 3 1 2 6 7 8 9 10 0 0,5 1 1,5 2 km
Tuulivoimala Rakennukset Asuinrakennus Liikerak. tai Julkinen rak. Lomarakennus Teollinen rakennus Kirkollinen rakennus Varjostus "real case" h/a 1 h/a 8 h/a 20 h/a 4 5 3 1 2 6 7 8 9 10 0 0,5 1 1,5 2 km
Bounds on non-surjective cellular automata
 Bounds on non-surjective cellular automata Jarkko Kari Pascal Vanier Thomas Zeume University of Turku LIF Marseille Universität Hannover 27 august 2009 J. Kari, P. Vanier, T. Zeume (UTU) Bounds on non-surjective
Bounds on non-surjective cellular automata Jarkko Kari Pascal Vanier Thomas Zeume University of Turku LIF Marseille Universität Hannover 27 august 2009 J. Kari, P. Vanier, T. Zeume (UTU) Bounds on non-surjective
Supplementary Information
 Supplementary Information Supplementary Table 1. Strain table Strain name Genotype Reference CEN.PK111-27B MATa leu2 trp1 Euroscarf TC-49 TC-50 pdc5::aro4* aro10::aro7* This study TC-50 MATa leu2 trp1
Supplementary Information Supplementary Table 1. Strain table Strain name Genotype Reference CEN.PK111-27B MATa leu2 trp1 Euroscarf TC-49 TC-50 pdc5::aro4* aro10::aro7* This study TC-50 MATa leu2 trp1
LX 70. Ominaisuuksien mittaustulokset 1-kerroksinen 2-kerroksinen. Fyysiset ominaisuudet, nimellisarvot. Kalvon ominaisuudet
 LX 70 % Läpäisy 36 32 % Absorptio 30 40 % Heijastus 34 28 % Läpäisy 72 65 % Heijastus ulkopuoli 9 16 % Heijastus sisäpuoli 9 13 Emissiivisyys.77.77 Auringonsuojakerroin.54.58 Auringonsäteilyn lämmönsiirtokerroin.47.50
LX 70 % Läpäisy 36 32 % Absorptio 30 40 % Heijastus 34 28 % Läpäisy 72 65 % Heijastus ulkopuoli 9 16 % Heijastus sisäpuoli 9 13 Emissiivisyys.77.77 Auringonsuojakerroin.54.58 Auringonsäteilyn lämmönsiirtokerroin.47.50
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
WindPRO version joulu 2012 Printed/Page :42 / 1. SHADOW - Main Result
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table 13.6.2013 19:42 / 1 Minimum
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table 13.6.2013 19:42 / 1 Minimum
Plasmid construction PCR reactions were carried out with Pfu or Pfu turbo polymerases (Stratagene) unless otherwise stated.
 Expanding the Genetic Code of Yeast for Incorporation of Diverse Unnatural Amino Acids via a Pyrrolysyl-tRNA Synthetase/tRNA Pair Susan M. Hancock 1, Rajendra Uprety 2, Alexander Deiters 2 & Jason W. Chin
Expanding the Genetic Code of Yeast for Incorporation of Diverse Unnatural Amino Acids via a Pyrrolysyl-tRNA Synthetase/tRNA Pair Susan M. Hancock 1, Rajendra Uprety 2, Alexander Deiters 2 & Jason W. Chin
KONEISTUSKOKOONPANON TEKEMINEN NX10-YMPÄRISTÖSSÄ
 KONEISTUSKOKOONPANON TEKEMINEN NX10-YMPÄRISTÖSSÄ https://community.plm.automation.siemens.com/t5/tech-tips- Knowledge-Base-NX/How-to-simulate-any-G-code-file-in-NX- CAM/ta-p/3340 Koneistusympäristön määrittely
KONEISTUSKOKOONPANON TEKEMINEN NX10-YMPÄRISTÖSSÄ https://community.plm.automation.siemens.com/t5/tech-tips- Knowledge-Base-NX/How-to-simulate-any-G-code-file-in-NX- CAM/ta-p/3340 Koneistusympäristön määrittely
,0 Yes ,0 120, ,8
 SHADOW - Main Result Calculation: Alue 2 ( x 9 x HH120) TuuliSaimaa kaavaluonnos Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered
SHADOW - Main Result Calculation: Alue 2 ( x 9 x HH120) TuuliSaimaa kaavaluonnos Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered
Network to Get Work. Tehtäviä opiskelijoille Assignments for students. www.laurea.fi
 Network to Get Work Tehtäviä opiskelijoille Assignments for students www.laurea.fi Ohje henkilöstölle Instructions for Staff Seuraavassa on esitetty joukko tehtäviä, joista voit valita opiskelijaryhmällesi
Network to Get Work Tehtäviä opiskelijoille Assignments for students www.laurea.fi Ohje henkilöstölle Instructions for Staff Seuraavassa on esitetty joukko tehtäviä, joista voit valita opiskelijaryhmällesi
Characterization of clay using x-ray and neutron scattering at the University of Helsinki and ILL
 Characterization of clay using x-ray and neutron scattering at the University of Helsinki and ILL Ville Liljeström, Micha Matusewicz, Kari Pirkkalainen, Jussi-Petteri Suuronen and Ritva Serimaa 13.3.2012
Characterization of clay using x-ray and neutron scattering at the University of Helsinki and ILL Ville Liljeström, Micha Matusewicz, Kari Pirkkalainen, Jussi-Petteri Suuronen and Ritva Serimaa 13.3.2012
Olet vastuussa osaamisestasi
 Olet vastuussa osaamisestasi Ohjelmistoammattilaisuuden uudet haasteet Timo Vehmaro 02-12-2015 1 Nokia 2015 Mitä osaamista tulevaisuudessa tarvitaan? Vahva perusosaaminen on kaiken perusta Implementaatio
Olet vastuussa osaamisestasi Ohjelmistoammattilaisuuden uudet haasteet Timo Vehmaro 02-12-2015 1 Nokia 2015 Mitä osaamista tulevaisuudessa tarvitaan? Vahva perusosaaminen on kaiken perusta Implementaatio
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
Constructive Alignment in Specialisation Studies in Industrial Pharmacy in Finland
 Constructive Alignment in Specialisation Studies in Industrial Pharmacy in Finland Anne Mari Juppo, Nina Katajavuori University of Helsinki Faculty of Pharmacy 23.7.2012 1 Background Pedagogic research
Constructive Alignment in Specialisation Studies in Industrial Pharmacy in Finland Anne Mari Juppo, Nina Katajavuori University of Helsinki Faculty of Pharmacy 23.7.2012 1 Background Pedagogic research
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
Security server v6 installation requirements
 CSC Security server v6 installation requirements Security server version 6.4-0-201505291153 Pekka Muhonen 8/12/2015 Date Version Description 18.12.2014 0.1 Initial version 10.02.2015 0.2 Major changes
CSC Security server v6 installation requirements Security server version 6.4-0-201505291153 Pekka Muhonen 8/12/2015 Date Version Description 18.12.2014 0.1 Initial version 10.02.2015 0.2 Major changes
LYTH-CONS CONSISTENCY TRANSMITTER
 LYTH-CONS CONSISTENCY TRANSMITTER LYTH-INSTRUMENT OY has generate new consistency transmitter with blade-system to meet high technical requirements in Pulp&Paper industries. Insurmountable advantages are
LYTH-CONS CONSISTENCY TRANSMITTER LYTH-INSTRUMENT OY has generate new consistency transmitter with blade-system to meet high technical requirements in Pulp&Paper industries. Insurmountable advantages are
Choose Finland-Helsinki Valitse Finland-Helsinki
 Write down the Temporary Application ID. If you do not manage to complete the form you can continue where you stopped with this ID no. Muista Temporary Application ID. Jos et onnistu täyttää lomake loppuun
Write down the Temporary Application ID. If you do not manage to complete the form you can continue where you stopped with this ID no. Muista Temporary Application ID. Jos et onnistu täyttää lomake loppuun
Infrastruktuurin asemoituminen kansalliseen ja kansainväliseen kenttään Outi Ala-Honkola Tiedeasiantuntija
 Infrastruktuurin asemoituminen kansalliseen ja kansainväliseen kenttään Outi Ala-Honkola Tiedeasiantuntija 1 Asemoitumisen kuvaus Hakemukset parantuneet viime vuodesta, mutta paneeli toivoi edelleen asemoitumisen
Infrastruktuurin asemoituminen kansalliseen ja kansainväliseen kenttään Outi Ala-Honkola Tiedeasiantuntija 1 Asemoitumisen kuvaus Hakemukset parantuneet viime vuodesta, mutta paneeli toivoi edelleen asemoitumisen
The Viking Battle - Part Version: Finnish
 The Viking Battle - Part 1 015 Version: Finnish Tehtävä 1 Olkoon kokonaisluku, ja olkoon A n joukko A n = { n k k Z, 0 k < n}. Selvitä suurin kokonaisluku M n, jota ei voi kirjoittaa yhden tai useamman
The Viking Battle - Part 1 015 Version: Finnish Tehtävä 1 Olkoon kokonaisluku, ja olkoon A n joukko A n = { n k k Z, 0 k < n}. Selvitä suurin kokonaisluku M n, jota ei voi kirjoittaa yhden tai useamman
Security server v6 installation requirements
 CSC Security server v6 installation requirements Security server version 6.x. Version 0.2 Pekka Muhonen 2/10/2015 Date Version Description 18.12.2014 0.1 Initial version 10.02.2015 0.2 Major changes Contents
CSC Security server v6 installation requirements Security server version 6.x. Version 0.2 Pekka Muhonen 2/10/2015 Date Version Description 18.12.2014 0.1 Initial version 10.02.2015 0.2 Major changes Contents
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table 5.11.2013 16:44 / 1 Minimum
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table 5.11.2013 16:44 / 1 Minimum
HARJOITUS- PAKETTI A
 Logistiikka A35A00310 Tuotantotalouden perusteet HARJOITUS- PAKETTI A (6 pistettä) TUTA 19 Luento 3.Ennustaminen County General 1 piste The number of heart surgeries performed at County General Hospital
Logistiikka A35A00310 Tuotantotalouden perusteet HARJOITUS- PAKETTI A (6 pistettä) TUTA 19 Luento 3.Ennustaminen County General 1 piste The number of heart surgeries performed at County General Hospital
MUSEOT KULTTUURIPALVELUINA
 Elina Arola MUSEOT KULTTUURIPALVELUINA Tutkimuskohteena Mikkelin museot Opinnäytetyö Kulttuuripalvelujen koulutusohjelma Marraskuu 2005 KUVAILULEHTI Opinnäytetyön päivämäärä 25.11.2005 Tekijä(t) Elina
Elina Arola MUSEOT KULTTUURIPALVELUINA Tutkimuskohteena Mikkelin museot Opinnäytetyö Kulttuuripalvelujen koulutusohjelma Marraskuu 2005 KUVAILULEHTI Opinnäytetyön päivämäärä 25.11.2005 Tekijä(t) Elina
FinFamily PostgreSQL installation ( ) FinFamily PostgreSQL
 FinFamily PostgreSQL 1 Sisällys / Contents FinFamily PostgreSQL... 1 1. Asenna PostgreSQL tietokanta / Install PostgreSQL database... 3 1.1. PostgreSQL tietokannasta / About the PostgreSQL database...
FinFamily PostgreSQL 1 Sisällys / Contents FinFamily PostgreSQL... 1 1. Asenna PostgreSQL tietokanta / Install PostgreSQL database... 3 1.1. PostgreSQL tietokannasta / About the PostgreSQL database...
Results on the new polydrug use questions in the Finnish TDI data
 Results on the new polydrug use questions in the Finnish TDI data Multi-drug use, polydrug use and problematic polydrug use Martta Forsell, Finnish Focal Point 28/09/2015 Martta Forsell 1 28/09/2015 Esityksen
Results on the new polydrug use questions in the Finnish TDI data Multi-drug use, polydrug use and problematic polydrug use Martta Forsell, Finnish Focal Point 28/09/2015 Martta Forsell 1 28/09/2015 Esityksen
Pricing policy: The Finnish experience
 Pricing policy: The Finnish experience Esa Österberg Senior Researcher Alcohol and Drug Research, STAKES, Helsinki, Finland esa.osterberg@stakes.fi Three pillars of traditional Nordic alcohol control Strict
Pricing policy: The Finnish experience Esa Österberg Senior Researcher Alcohol and Drug Research, STAKES, Helsinki, Finland esa.osterberg@stakes.fi Three pillars of traditional Nordic alcohol control Strict
3 9-VUOTIAIDEN LASTEN SUORIUTUMINEN BOSTONIN NIMENTÄTESTISTÄ
 Puhe ja kieli, 27:4, 141 147 (2007) 3 9-VUOTIAIDEN LASTEN SUORIUTUMINEN BOSTONIN NIMENTÄTESTISTÄ Soile Loukusa, Oulun yliopisto, suomen kielen, informaatiotutkimuksen ja logopedian laitos & University
Puhe ja kieli, 27:4, 141 147 (2007) 3 9-VUOTIAIDEN LASTEN SUORIUTUMINEN BOSTONIN NIMENTÄTESTISTÄ Soile Loukusa, Oulun yliopisto, suomen kielen, informaatiotutkimuksen ja logopedian laitos & University
Rakennukset Varjostus "real case" h/a 0,5 1,5
 Tuulivoimala Rakennukset Asuinrakennus Liikerak. tai Julkinen rak. Lomarakennus Teollinen rakennus Kirkollinen rakennus Varjostus "real case" h/a 1 h/a 8 h/a 20 h/a 1 2 3 5 8 4 6 7 9 10 0 0,5 1 1,5 2 km
Tuulivoimala Rakennukset Asuinrakennus Liikerak. tai Julkinen rak. Lomarakennus Teollinen rakennus Kirkollinen rakennus Varjostus "real case" h/a 1 h/a 8 h/a 20 h/a 1 2 3 5 8 4 6 7 9 10 0 0,5 1 1,5 2 km
WindPRO version joulu 2012 Printed/Page :47 / 1. SHADOW - Main Result
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
lpar1 IPB004065, IPB002277, and IPB Restriction Enyzme Differences from REBASE Gained in Variant Lost from Reference
 lpar1 IPB465, IPB2277, and IPB5385 Genomic Sequence Coding Sequence For help interpreting these results, view the PARSENP Introduction page. # View On Sequence Nucleotide Change Effect Restriction Enyzme
lpar1 IPB465, IPB2277, and IPB5385 Genomic Sequence Coding Sequence For help interpreting these results, view the PARSENP Introduction page. # View On Sequence Nucleotide Change Effect Restriction Enyzme
Sisällysluettelo Table of contents
 Sisällysluettelo Table of contents OTC:n Moodlen käyttöohje suomeksi... 1 Kirjautuminen Moodleen... 2 Ensimmäinen kirjautuminen Moodleen... 2 Salasanan vaihto... 2 Oma käyttäjäprofiili... 3 Työskentely
Sisällysluettelo Table of contents OTC:n Moodlen käyttöohje suomeksi... 1 Kirjautuminen Moodleen... 2 Ensimmäinen kirjautuminen Moodleen... 2 Salasanan vaihto... 2 Oma käyttäjäprofiili... 3 Työskentely
ELEMET- MOCASTRO. Effect of grain size on A 3 temperatures in C-Mn and low alloyed steels - Gleeble tests and predictions. Period
 1 ELEMET- MOCASTRO Effect of grain size on A 3 temperatures in C-Mn and low alloyed steels - Gleeble tests and predictions Period 20.02-25.05.2012 Diaarinumero Rahoituspäätöksen numero 1114/31/2010 502/10
1 ELEMET- MOCASTRO Effect of grain size on A 3 temperatures in C-Mn and low alloyed steels - Gleeble tests and predictions Period 20.02-25.05.2012 Diaarinumero Rahoituspäätöksen numero 1114/31/2010 502/10
On instrument costs in decentralized macroeconomic decision making (Helsingin Kauppakorkeakoulun julkaisuja ; D-31)
 On instrument costs in decentralized macroeconomic decision making (Helsingin Kauppakorkeakoulun julkaisuja ; D-31) Juha Kahkonen Click here if your download doesn"t start automatically On instrument costs
On instrument costs in decentralized macroeconomic decision making (Helsingin Kauppakorkeakoulun julkaisuja ; D-31) Juha Kahkonen Click here if your download doesn"t start automatically On instrument costs
rapid evolution in copy number, location and sequence, with diverse turnover mechanisms.
 Repetitive DNA and its evolution: genomic strategies to understand the processes and exploit biodiversity rapid evolution in copy number, location and sequence, with diverse turnover mechanisms. often
Repetitive DNA and its evolution: genomic strategies to understand the processes and exploit biodiversity rapid evolution in copy number, location and sequence, with diverse turnover mechanisms. often
7.4 Variability management
 7.4 Variability management time... space software product-line should support variability in space (different products) support variability in time (maintenance, evolution) 1 Product variation Product
7.4 Variability management time... space software product-line should support variability in space (different products) support variability in time (maintenance, evolution) 1 Product variation Product
National Building Code of Finland, Part D1, Building Water Supply and Sewerage Systems, Regulations and guidelines 2007
 National Building Code of Finland, Part D1, Building Water Supply and Sewerage Systems, Regulations and guidelines 2007 Chapter 2.4 Jukka Räisä 1 WATER PIPES PLACEMENT 2.4.1 Regulation Water pipe and its
National Building Code of Finland, Part D1, Building Water Supply and Sewerage Systems, Regulations and guidelines 2007 Chapter 2.4 Jukka Räisä 1 WATER PIPES PLACEMENT 2.4.1 Regulation Water pipe and its
Python Libraries 1 / 14
 Python Libraries 1 / 14 Gzip, CSV https://docs.python.org/3/library/gzip.html https://docs.python.org/3/library/csv.html https://daler.github.io/pybedtools/ - PyBed tools http://biopython.org/wiki/gff_parsing
Python Libraries 1 / 14 Gzip, CSV https://docs.python.org/3/library/gzip.html https://docs.python.org/3/library/csv.html https://daler.github.io/pybedtools/ - PyBed tools http://biopython.org/wiki/gff_parsing
Bioinformatics. Sequence Analysis: Part III. Pattern Searching and Gene Finding. Fran Lewitter, Ph.D. Head, Biocomputing Whitehead Institute
 Bioinformatics Sequence Analysis: Part III. Pattern Searching and Gene Finding Fran Lewitter, Ph.D. Head, Biocomputing Whitehead Institute Course Syllabus Jan 7 Jan 14 Jan 21 Jan 28 Feb 4 Feb 11 Feb 18
Bioinformatics Sequence Analysis: Part III. Pattern Searching and Gene Finding Fran Lewitter, Ph.D. Head, Biocomputing Whitehead Institute Course Syllabus Jan 7 Jan 14 Jan 21 Jan 28 Feb 4 Feb 11 Feb 18
TEST REPORT Nro VTT-S Air tightness and strength tests for Furanflex exhaust air ducts
 TEST REPORT Nro VTT-S-04515-08 19.5.008 Air tightness and strength tests for Furanflex exhaust air ducts Requested by: Hormex Oy TEST REPORT NRO VTT-S-04515-08 1 () Requested by Order Hormex Oy Linnanherrankuja
TEST REPORT Nro VTT-S-04515-08 19.5.008 Air tightness and strength tests for Furanflex exhaust air ducts Requested by: Hormex Oy TEST REPORT NRO VTT-S-04515-08 1 () Requested by Order Hormex Oy Linnanherrankuja
( N117 x HH141 ( Honkajoki N117 x 9 x HH120 tv-alueet ( ( ( ( ( ( ( ( ( ( m. Honkajoki & Kankaanpää tuulivoimahankkeet
 Honkajoki & Kankaanpää tuulivoimahankkeet N117 x HH141 Honkajoki N117 x 9 x HH120 tv-alueet Alahonkajoki_kaava_alueen_raja_polyline Asuinrakennus Julkinen tai liiker rak. Lomarakennus Teollinen rak. Allas
Honkajoki & Kankaanpää tuulivoimahankkeet N117 x HH141 Honkajoki N117 x 9 x HH120 tv-alueet Alahonkajoki_kaava_alueen_raja_polyline Asuinrakennus Julkinen tai liiker rak. Lomarakennus Teollinen rak. Allas
Genome 373: Genomic Informatics. Professors Elhanan Borenstein and Jay Shendure
 Genome 373: Genomic Informatics Professors Elhanan Borenstein and Jay Shendure Genome 373 This course is intended to introduce students to the breadth of problems and methods in computational analysis
Genome 373: Genomic Informatics Professors Elhanan Borenstein and Jay Shendure Genome 373 This course is intended to introduce students to the breadth of problems and methods in computational analysis
VIIKKI BIOCENTER University of Helsinki
 VIIKKI BIOCENTER University of Helsinki Mitä uudet DNA sekvensointimenetelmät voivat paljastaa luonnonjärjestelmästä? Petri Auvinen DNA Sequencing and Genomics Laboratory Institute of Biotechnology Petri
VIIKKI BIOCENTER University of Helsinki Mitä uudet DNA sekvensointimenetelmät voivat paljastaa luonnonjärjestelmästä? Petri Auvinen DNA Sequencing and Genomics Laboratory Institute of Biotechnology Petri
Valuation of Asian Quanto- Basket Options
 Valuation of Asian Quanto- Basket Options (Final Presentation) 21.11.2011 Thesis Instructor and Supervisor: Prof. Ahti Salo Työn saa tallentaa ja julkistaa Aalto-yliopiston avoimilla verkkosivuilla. Muilta
Valuation of Asian Quanto- Basket Options (Final Presentation) 21.11.2011 Thesis Instructor and Supervisor: Prof. Ahti Salo Työn saa tallentaa ja julkistaa Aalto-yliopiston avoimilla verkkosivuilla. Muilta
Strain or plasmid Description a Reference or source b. 168 trpc2 Laboratory. 1A780 trpc2 sigb::spc BGSC
 TABLE S1 Bacterial strains and plasmids Strain or plasmid Description a Reference or source b B. subtilis 168 trpc2 Laboratory stock 1A780 trpc2 sigb::spc BGSC BM1010 trpc2 htpx::pgs1582; Em r BM1302 trpc2
TABLE S1 Bacterial strains and plasmids Strain or plasmid Description a Reference or source b B. subtilis 168 trpc2 Laboratory stock 1A780 trpc2 sigb::spc BGSC BM1010 trpc2 htpx::pgs1582; Em r BM1302 trpc2
Guideline on Similar biological medicinal products containing biotechnology-derived proteins as active substance: non-clinical and clinical issues
 Guideline on Similar biological medicinal products containing biotechnology-derived proteins as active substance: non-clinical and clinical issues EMA Workshop on Biosimilars, 31 October 2014 Pekka Kurki
Guideline on Similar biological medicinal products containing biotechnology-derived proteins as active substance: non-clinical and clinical issues EMA Workshop on Biosimilars, 31 October 2014 Pekka Kurki
Lab SBS3.FARM_Hyper-V - Navigating a SharePoint site
 Lab SBS3.FARM_Hyper-V - Navigating a SharePoint site Note! Before starting download and install a fresh version of OfficeProfessionalPlus_x64_en-us. The instructions are in the beginning of the exercise.
Lab SBS3.FARM_Hyper-V - Navigating a SharePoint site Note! Before starting download and install a fresh version of OfficeProfessionalPlus_x64_en-us. The instructions are in the beginning of the exercise.
Tilausvahvistus. Anttolan Urheilijat HENNA-RIIKKA HAIKONEN KUMMANNIEMENTIE 5 B RAHULA. Anttolan Urheilijat
 7.80.4 Asiakasnumero: 3000359 KALLE MANNINEN KOVASTENLUODONTIE 46 51600 HAUKIVUORI Toimitusosoite: KUMMANNIEMENTIE 5 B 51720 RAHULA Viitteenne: Henna-Riikka Haikonen Viitteemme: Pyry Niemi +358400874498
7.80.4 Asiakasnumero: 3000359 KALLE MANNINEN KOVASTENLUODONTIE 46 51600 HAUKIVUORI Toimitusosoite: KUMMANNIEMENTIE 5 B 51720 RAHULA Viitteenne: Henna-Riikka Haikonen Viitteemme: Pyry Niemi +358400874498
Asiakaspalautteen merkitys laboratoriovirheiden paljastamisessa. Taustaa
 Asiakaspalautteen merkitys laboratoriovirheiden paljastamisessa Paula Oja, TtT Laboratorio, Oulun yliopistollinen sairaala Potilasturvallisuustutkimuksen päivät 26. 27.1.2011 1 Taustaa Laboratorion tulee
Asiakaspalautteen merkitys laboratoriovirheiden paljastamisessa Paula Oja, TtT Laboratorio, Oulun yliopistollinen sairaala Potilasturvallisuustutkimuksen päivät 26. 27.1.2011 1 Taustaa Laboratorion tulee
I. Principles of Pointer Year Analysis
 I. Principles of Pointer Year Analysis Fig 1. Maximum (red) and minimum (blue) pointer years. 1 Fig 2. Principle of pointer year calculation. Fig 3. Skeleton plot graph created by Kinsys/Kigraph programme.
I. Principles of Pointer Year Analysis Fig 1. Maximum (red) and minimum (blue) pointer years. 1 Fig 2. Principle of pointer year calculation. Fig 3. Skeleton plot graph created by Kinsys/Kigraph programme.
 Digitally signed by Hans Vadbäck DN: cn=hans Vadbäck, o, ou=fcg Suunnittelu ja Tekniikka Oy, email=hans.vadback@fcg.fi, c=fi Date: 2016.12.20 15:45:35 +02'00' Jakob Kjellman Digitally signed by Jakob Kjellman
Digitally signed by Hans Vadbäck DN: cn=hans Vadbäck, o, ou=fcg Suunnittelu ja Tekniikka Oy, email=hans.vadback@fcg.fi, c=fi Date: 2016.12.20 15:45:35 +02'00' Jakob Kjellman Digitally signed by Jakob Kjellman
Predicting evolutionarily conserved regions (ECRs) in the Xenopus tropicalis genome using a MultiPipMaker-based bioinformatic strategy
 Predicting evolutionarily conserved regions (ECRs) in the Xenopus tropicalis genome using a MultiPipMaker-based bioinformatic strategy OVERVIEW- Method updated and written by Sarah Louie (6/19/08) Initially
Predicting evolutionarily conserved regions (ECRs) in the Xenopus tropicalis genome using a MultiPipMaker-based bioinformatic strategy OVERVIEW- Method updated and written by Sarah Louie (6/19/08) Initially
FinFamily Installation and importing data (11.1.2016) FinFamily Asennus / Installation
 FinFamily Asennus / Installation 1 Sisällys / Contents FinFamily Asennus / Installation... 1 1. Asennus ja tietojen tuonti / Installation and importing data... 4 1.1. Asenna Java / Install Java... 4 1.2.
FinFamily Asennus / Installation 1 Sisällys / Contents FinFamily Asennus / Installation... 1 1. Asennus ja tietojen tuonti / Installation and importing data... 4 1.1. Asenna Java / Install Java... 4 1.2.
Kvanttilaskenta - 1. tehtävät
 Kvanttilaskenta -. tehtävät Johannes Verwijnen January 9, 0 edx-tehtävät Vastauksissa on käytetty edx-kurssin materiaalia.. Problem False, sillä 0 0. Problem False, sillä 0 0 0 0. Problem A quantum state
Kvanttilaskenta -. tehtävät Johannes Verwijnen January 9, 0 edx-tehtävät Vastauksissa on käytetty edx-kurssin materiaalia.. Problem False, sillä 0 0. Problem False, sillä 0 0 0 0. Problem A quantum state
FYSE301(Elektroniikka(1(A3osa,(kevät(2013(
 FYSE301(Elektroniikka(1(A3osa,(kevät(2013( 1/2 Loppukoe1.3.2013 vastaakaikkiinkysymyksiin(yhteensä48pistettä) 1. Kuvailelyhyesti a. Energialineaarisissapiirielementeissä:vastuksessa,kondensaattorissajakelassa(3
FYSE301(Elektroniikka(1(A3osa,(kevät(2013( 1/2 Loppukoe1.3.2013 vastaakaikkiinkysymyksiin(yhteensä48pistettä) 1. Kuvailelyhyesti a. Energialineaarisissapiirielementeissä:vastuksessa,kondensaattorissajakelassa(3
Accommodation statistics
 Transport and Tourism 2011 Accommodation statistics 2011, January Nights spent by foreign tourists in Finland increased by per cent in January The number of recorded nights spent by foreign tourists at
Transport and Tourism 2011 Accommodation statistics 2011, January Nights spent by foreign tourists in Finland increased by per cent in January The number of recorded nights spent by foreign tourists at
Ajettavat luokat: SM: S1 (25 aika-ajon nopeinta)
 SUPERMOTO SM 2013 OULU Lisämääräys ja ohje Oulun Moottorikerho ry ja Oulun Formula K-125ry toivottaa SuperMoto kuljettajat osallistumaan SuperMoto SM 2013 Oulu osakilpailuun. Kilpailu ajetaan karting radalla
SUPERMOTO SM 2013 OULU Lisämääräys ja ohje Oulun Moottorikerho ry ja Oulun Formula K-125ry toivottaa SuperMoto kuljettajat osallistumaan SuperMoto SM 2013 Oulu osakilpailuun. Kilpailu ajetaan karting radalla
Increase of opioid use in Finland when is there enough key indicator data to state a trend?
 Increase of opioid use in Finland when is there enough key indicator data to state a trend? Martta Forsell, Finnish Focal Point 28.9.2015 Esityksen nimi / Tekijä 1 Martta Forsell Master of Social Sciences
Increase of opioid use in Finland when is there enough key indicator data to state a trend? Martta Forsell, Finnish Focal Point 28.9.2015 Esityksen nimi / Tekijä 1 Martta Forsell Master of Social Sciences
Solar Water Heater Kit. EcoStyle. 1 User Manual/Operating Instructions. Contents FREE LESSON PLANS AVAILABLE.
 EcoStyle 1 User Manual/Operating Instructions Contents 1 Contents 2 What s in the box? 3 Setting up the 4 Heating water 5 Measuring water temperature with the digital thermometer 6 Maximum/minimum temperature
EcoStyle 1 User Manual/Operating Instructions Contents 1 Contents 2 What s in the box? 3 Setting up the 4 Heating water 5 Measuring water temperature with the digital thermometer 6 Maximum/minimum temperature
MIKES, Julkaisu J3/2000 MASS COMPARISON M3. Comparison of 1 kg and 10 kg weights between MIKES and three FINAS accredited calibration laboratories
 MITTATEKNIIKAN KESKUS CENTRE FOR METROLOGY AND ACCREDITATION Julkaisu J3/2000 MASS COMPARISON M3 Comparison of 1 kg and 10 kg weights between MIKES and three FINAS accredited calibration laboratories Kari
MITTATEKNIIKAN KESKUS CENTRE FOR METROLOGY AND ACCREDITATION Julkaisu J3/2000 MASS COMPARISON M3 Comparison of 1 kg and 10 kg weights between MIKES and three FINAS accredited calibration laboratories Kari
Capacity utilization
 Mat-2.4142 Seminar on optimization Capacity utilization 12.12.2007 Contents Summary of chapter 14 Related DEA-solver models Illustrative examples Measure of technical capacity utilization Price-based measure
Mat-2.4142 Seminar on optimization Capacity utilization 12.12.2007 Contents Summary of chapter 14 Related DEA-solver models Illustrative examples Measure of technical capacity utilization Price-based measure
Salasanan vaihto uuteen / How to change password
 Salasanan vaihto uuteen / How to change password Sisällys Salasanakäytäntö / Password policy... 2 Salasanan vaihto verkkosivulla / Change password on website... 3 Salasanan vaihto matkapuhelimella / Change
Salasanan vaihto uuteen / How to change password Sisällys Salasanakäytäntö / Password policy... 2 Salasanan vaihto verkkosivulla / Change password on website... 3 Salasanan vaihto matkapuhelimella / Change
WP3 Decision Support Technologies
 WP3 Decision Support Technologies 1 WP3 Decision Support Technologies WP Leader: Jarmo Laitinen Proposed budget: 185 000, VTT 100 000, TUT 85 000. WP3 focuses in utilizing decision support technologies
WP3 Decision Support Technologies 1 WP3 Decision Support Technologies WP Leader: Jarmo Laitinen Proposed budget: 185 000, VTT 100 000, TUT 85 000. WP3 focuses in utilizing decision support technologies
