Figure S1. CLUSTAL O(1.2.1) multiple sequence alignment
|
|
|
- Olivia Tamminen
- 6 vuotta sitten
- Katselukertoja:
Transkriptio
1 Figure S1. A CLUSTAL O(1.2.1) multiple sequence alignment AT5G MNVANPNEYPDMNPNAAQNRNMSRPDQQ 28 AT5G MANRPHVPKFGDWTEDAPFTVVFEKASKSKKNMNVANPNEYPDMNPNAAQNRNMSRPDQQ 60 **************************** AT5G PPNHNVRPRHERFNSRDETEFRPSPAHNERNNRVRSVPPTPETYNHQTYGGGGRSMGNPT 88 AT5G PPNHNVRPRHERFNSRDETEFRPSPAHNERNNRVRSVPPTPETYNHQTYGGGGRSMGNPT 120 ************************************************************ AT5G EINRRQSRDHVPARPIRNLRGQSSERVATIPPFPGTGSNMENQSYTLIFDKVKEDRNQAR 148 AT5G EINRRQSRDHVPARPIRNLRGQSSERVATIPPFPGTGSNMENQSYTLIFDKVKEDRNQAR 180 ************************************************************ AT5G SYNGTDHSTPTRPIIDQHHQPLPSSPKVDNQNIISLSFINPLFLQLTLLCVGLLFSSMEP 208 AT5G SYNGTDHSTPTRPIIDQHHQPLPSSPKGCCFPPWS R 216 *************************** * AT5G KGKLMKMIISLFYIILFFFICFCLIFFNNIFVILMEM 245 AT5G KGS **. B CLUSTAL O(1.2.1) multiple sequence alignment NOI1 GKPLPKFGEWDV 12 NOI2 DRPLPKFGEWDV 12 NOI3 GRALPKFGEWDV 12 NOI4 GRPLPKFGEWDV 12 NOI5 ARPLPKFGEWDA 12 NOI6 WLSVPQFGDWDQ 12 NOI7 WLSVPQFGDWDQ 12 NOI8 SGQIPRFGEWEE 12 NOI9 RGHVPAFGSWDW 12 NOI11 RPHVPKFGDWNN 12 NOI12 GRPLPKFGEWDV 12 NOI13 ARPLPKFGEWDV 12 NOI14 WTPVPQFGGWDQ 12 RIN4-RCS1 RSNVPKFGNWEA 12 RIN4-RCS2 VTVVPKFGDWDE 12 :* ** *: Figure S1. Alignments of both NOI10 gene models and of the predicted AvrRpt2 cleavage sites of all the NOI proteins. (A,B) Multiple sequence alignments were performed with CLUSTAL O(1.2.1) ( Sievers et al., 2011). (A) Sequence alignment of both NOI10 (AT5G48657) splice variants, the conserved AvrRpt2 cleavage site is highlighted in grey. (B) Sequence alignment of AvrRpt2 cleavage sites of RIN4/NOI protein family members. Note: the representative gene model of NOI10 (AT5G ) does not contain a cleavage site. 1
2 Figure S2. A 2
3 B ATG - start codon GCG - exon gta - intron TAA - stop codon cta UTR NOI1 At5g63270: tagggatttacgcgggaaagaagaagaggtgtaataatgttgtttgatttggttaatgc Insertion (SAIL_1173_C11) tgttgtcaacgacgacgacgacgtgtttccctcttatgtttctccttcctctctctatattacaactg catttcccttctctctcctcccacctccctctctctctctcaatagatctttgatatttcattttgct ttgatctcaacaacaatggcggtaagaacaaacaacgtcttcatcaagtctctctcctttcatcatca ccacgatgatcagtttcagaaaaagtctctatagctttcttgatgtttgatgaatctctgtttctttg gtgcaggaaaacaaagggaaaccattgccaaagtttggggaatgggatgtgaacaatccggcatcagc CGAAGGATTCACAGTCATTTTCAGCAAAGCTAGTGACGAGAAGAAGACCAAGAAAGCATCCGGCGCTG GCCCTAATAGTCTGGTTTCACCTCAGAGAAACCAAAACTCCGATCAAAACAATAACCATAGTTCCCAA AACCCTAAAGCTAAGgttgttttctttttctcattacattcaacttcttagctctttatccaagaaca tagatttgctcacctgatttatgctttctgcgcagaacaaatggttttgcttccgttaaatcctgcca tcttctgaagaatccgaggggtagggaagctgaagcagttttgcagagaggtctcaaacaaagtgaaa caaaagggttaacgcttaaagacccatatatctcagctttcttcttcttctctcttctgctgatatta tacttgttcagtatactttttgttcctttctttttgtttgccaacattttgcgcacaatggatataat ttgtatcttttgactctccatctaagaactatggaaataagattttgagtaattgtccttcgcagaca taatgctattagctattattcatatcgacatggctttctgtacatcgttccaatagct NOI4 At5g : agtgattcacgtcacttgggtctaataactccgtttatataaaccgttacagacatcgcacaccgaga gagagataataaagagagaagcaaaaaagaatttttatagaaagaaaggaaaaatcagaaaaggagag aaaaattaaaaaagtctctcttttctctttttcttcttcgtctcccatattctccgatcaaaaatata tttctctctcactttccttggttttctcggcggcttcttcttctcctatttctccctcaaatcgtttt ctccagttttcttgggataaatcttcctcctttctgcagacgccatgtcggtgcgtttaaatctcctc tctttattatatacatatgcttgagaacagttttgaattcgatttgtttggttttgagaattttagtt TCGTTTTGGTGAGTTAGATGATTATATGTTTCCTTTTGATTGGAGGgtttttgttgtttcttttatat ccgctgaaaatcttagattaaatagaaaaacagatgtgaaattgtgattctgatgagctattgtcttg ttgttgaatctgataatcctgattctggatctcgggttttattcaagttttgatctttttctgaattt gattgattaagcttagatttttgattcagggtttctgattttggatttgagatgaaataaaggtttga taaagagaagttgttttgttgtctttgtgttagcttatctttttttccccagattgttgttctttgat tttattgatagtgaaagaggcatgaatattgtgtggcattgtttatggacaagcaaaagttctttttg ccttgtggatttgtttgcttatttagagacacaattgaaatccactagtttcaaacttttggttttat tattttctcacctaatcaaatggattataaagttgtttggaactttaaaatttctcttatggtttggt tttggtaatccttttgacttgtgctctatattgtttgctacaaaatggaattataattagtttgggca aattgtacaaaccattttttcctcgttattgatgatagaaaaggttccattccttaagtcctcattct tatttacttggttgattctttttttcggtttatccttgcctcttctcttttgtatgtgctcccactta agacttgcttagcttatgaattcatctacttggcaagattgaacttatttcttgatttcaccatttgg tttttatatgaacaacaggacaaaggtcgtcccttgccaaaatttggtgaatgggatgtgaatgatcc AGCATCAGCAGAAGGTTTTACAGTGATATTCAACAAAGCTAGGGATGAGAAAAAGACCGGTGGCAAAC CGGGATCACCCGGTAAATCCAGTGAGGGTCATGTTAAATCTGGAGGAGGAGATCCTAGTAAACCTCAG CCTGTAAgtttcctcttcttcatctttatctccttgatccattgtcacactaaatggggttttatctt tggtttcagaaaaaatggctctgctgcatgcaagctccagctgtggactcttgacagacacaaagatg GATTTGCTTGCTGCTAAAAAAAAAGTCACAGTGCTTACACATCTAAAAGCAATGGTTCTTTTTATGTT TTATTGTCGTCTTTCTTGA Insertion (SALK_042784C) 3
4 aattacccacacacacaaaaaaaaaaggttcctaagatgatttggagctcacccacctttagtatcac gattatgattccttcttctttgaagttgtttcttcttcttaaaatgctgtaaaatcgtgtctccattt tattggaaaaaaaaaaggaagaaaaacgaaaaaaaacacagttactgtgtctatatgtaatgattctc ttggggtctttttgttttttgattttatggtgtctttttccttgattttttaaattatattatcttgg gctcctttttaatattttcctccttagtaaatcaaacaagtttcattaatgaaaacgaacaagaactt NOI5 At3g48450: atattacaatttcacctctctttttcccagatcatcatcatcctctctctttctctagattttggata tttcctcgtatcggattcgaaatcctctgtttctaattccaacaatggcaacggtaagaacaatatgc atctcttatatctacgaattctgattcatcttacatttcttgatatgatttcaaaacttccctccttt catacaattttccaaatctcttatcttgaaaaaaagtctcaaactttggctagaaatctgcatttttt ttaatgatgaacaattctgttttttgttttgtttgtttggtggtgtgtagggaaatagagcgagacca TTGCCAAAATTTGGGGAATGGGATGCGACTAATCCTGGCTCGGCTGAAGGATTCACTGTCATTTTCAA TAAAGCTCGCGATGACAAGAAGACGATGAAAACCGCTGTCGCAGGACCTGAGAGTATTGTTTCACCTC CAAGAAATGAAGAACCTCCTAAAAACAACAACAACCATCATCACAACCATCACACCCGTCATTCCCAA ACCCCAAGATCAAAGgtacttttttgccacagatactcaaagggtatatattaggtatttctctgctt gacttgttatgtttttgttcttttgttgtgcagaagaaatggctttgttttcgttaactctggtattg Insertion (SALK_118446) gctctgttctgagaaaacccgacgacaagcaagcagaatcgtggagcgagtaatctccaacagattat agcgaccaaaccagtgatcatctaagatgttgttgcaaacttctcttcttcttattgctcgttgagtt tctttctcttttgttttggacacatttgggtatatgaatatacgtttttcttgatgattcttagtttc ataagtatatataaag NOI7 At5g09960: tagtgattgctgattgggtataattcgagctgtttttttgctgtaataatttcacaattctctttttt tttcgcttttaaataattttgtctctccatcttcttctcttttgctagtctctcatatcagctaagaa aagaaattcagaacaaaaaaataacacaaagctctgtgtttctgtctatctgttgaatcaaatcatat GGAAGACGATCGAAAAGAGgtataaccatctgttttattatctcttgttagtttccccataaagtttc tttaggaaactttgattttactaagaaatcgagatctgcttgtttggattttttcgagaattgaacat tggttttgtagagctaatcaagctccaagcactactgatactgtttatattgaagattttgattaaaa atcagcactaatcttggtaatcggaaacaaaaaaaattgagttagtagtatattctaaagtttcatcc tttttcacatctcgataagctgaaattttgttatcacgaggtatattttctactagtacttggtaagt tcgaaactttattggattgataaagaaaagatcttactttaccctgattttatcgatcattgattaaa ccaaagcttgtaaattgtcccataaagtcaagaacttgactttgtttcttagagagttcgtcctcttt cttcttcacttgcttgctcatgatgcatcgttttgtaaatagctctgtttttttttcctttaagtata tgcttcattatgatttatgaaacctttttaagtttggtaatgtttgtttcgaaatgtctttgtttttg cagaagaacactccgtggctatcagtgccacagtttggtgattgggaccaaaaaggaggaggaacaat GCCTGATTACTCTATGGATTTCACTAAGATTAGAGAGATGAGGAAACAAAACAAGAGAGACCCTTCTC GAGCCAGTTTAGGCAATGAGGAAGAGCTCATTAAGCCACCCGAGTCAGCAACATCAACTGCTGAGCTT ACCACGGTCCAAAGTGAAAACCAACGAGAGTTCTCTCCCAGCCACCATCATCAACCACATTCTCCTTC Tgtaagattcttcaccatt Insertion (SALK_007305C) atttggtttatatcatattattccttaattatatttgcattactatcattaagacatttagaggcata ttactatattagcacatggttttgtaatcttgcaatctatatacagtattaaaattttgaatagttgt atttgagctggtagtgattatcactgtggaatagatgagaatttggtgtggttgttttttttgacaga CGAGGAGAAGTATGTTCAGCTGCTTCAACTGCTGCGTTAAAGCTTGAagatttcttcttgagcaaagt agcagttttattattgacttgtgatttgaatgtggaaatgtgttaatgtcatgacactttaatatatg ttccaatccatttttctttttctttgggaaccatataagctctattacaaggaaaagattatggggga aattaagtatacacaaaaacataatcgtctctattagtacactct NOI8 At5g : 4
5 5 gactatttagacttttagaggtaaggacaatatcttctttgcttacataccccactcttgttataaaa gacagtaacctttcttgagatcactctttcttatttgtgtgttagtgcattgagagagagagagagag atagaaagagaaagaaatcgtcatggctcatgtaagtgaaacttctctaatcctctatccttgtttct tcctgtcgtctttaacatatagatcgtcaaaac Insertion (SALK_101426) ctgcctaataatgtcttatcctcacataacacctttttttttatctccatctctctgttacttagaac cctccattcatatatacactaggtgccaaatttggtcacgtgtttggcccttcacgttcttgactctt agctagatccttgtttcttgtggattcttgaatcttaaatcttgaatccctgagttttgttggttctt gattttgattctcgattctttagtcattgattattcgatctttgttggtttttgattagagatttaat tagcatatttatgtactaataattgaataaatgggaacaggggagatatgatacttacaagaagaaaa GTGGACAGATTCCTCGGTTTGGAGAATGGGAAGAAGCAAATGAGATGCCAATAACACAATACTTTGAG AATCCAAGACAAGCTGGTCTGATTCGTCACCAATACACAACAACTTCTTCTGCTTCTTCCACTACCAC TTCATCTTCATCTTCTTCTTCTTCTTCTGCAGAAGCCTTAAAGCTTGCTTCTCACCATCCCCGTCCAC GTCACCTCCACGCTCAAAGACAGgttcattcatctaatcactacttatttttcttaattatataacat ttatttaactcgataaacgtaattaagtatctaaagaagaacgcgttgtttgtttgattaatgacttg gcgtagacggcggggacgaaggagaaaagaggaccacaaaggcgtgtgcgtgacgtcagtgcacagtc GGACAAGTATTACATTGACGTCAACGGTGTTAAGCAGTTCAAAAACGACGTTGCTCTGACTTGTAAGC CACCTAAGCCCGTTGATGAAGATCTCTACAAGATTCCTCCTGAGTTTATCCATTCTTCAACAAGGgta agagctccttctcacacacaacttattttactgcattttataacaaagttttatatatttttaatcaa cttttcaattttatatcactttttctttaactttgtttgttatttaaaaagagttttaaaataaattt ggtgttgcagaagagaaggcctagctttttagcttgtttggttccatgcgcatgaatcgaaaaggaag agcgaagaagaagaagaaaaagtaggatcatgaatgaataatgtaatttgttttttgtttttgtaatt tcatcctaatatacagatatgatatatacgtggtttgacagaaaaagagagcatacgagtggtttttg aaaaagagtaacaaagtaatatcgtagtaaactagagtaatatatccttttgtcttgttacatatcta ctttataagattttctctcatttttttttcacgt NOI9 At5g48500: tatattattttcacttaccctacactatatttttgaaaacaccactctatgctcagagacacacagtc cctcactactctttctctatctcctacaagtagactttaagacaacttttgtctcagtgttgagtaat gtaaaatacccatctggttatttattacgcatagatctctctcttctctctttctctatttagattac ttaaagcagagaggtccgaacatcatcaatcaaacccattcacaactactacgacaatggactacgta agctccttcctctttcttcttcttcttcgtcatcttcttgtgggttatacaaagtttttaacttttta tgttttctcaatcaggaatacggaggaggagagtactgtcggagaggacac Insertion (SALK_150777C) GTGCCAGCTTTTGGAAGCTGGGATTGGAACGACGCCGTTCCATTCACTCAGTGTTTCGAGACAGCAAC AACTCAACAACCAGCTTTTCTTCACTATGCTTCTTACCCTCAAGATCGTGATCTTTACCTCGCCGGTG ATCTCTACGACAACCACCACCTTGTAGCTCCGGCCGTTATCATTGTCCCTCGTCGCCGGgtactttct tgattcatgtattcaacacgacgccttgagtttcttgatttagtcttatgttcgtggaaaatgtttat ttctaggctaaagtgggtcaagaaccagcggagaagaagagaaatgtgtcgaagcagcaacacaacta TAAGACGGAGGCGCGTGAGTTTAACGCGCCGGTAAGCTGTCCAACACCGGTTGTGAAGCGGAGGATGA AGGCACCAAAGCCTGTGGATGAAGACTTGTACAAAGTTTCCCCACAACTTCTCTCAGTCAAATCCAAA AGGgtaattaaactttaatttaattaacttgttattaaattagtcttatctttgttagtctgcttcga ttaatgattttgattatggaaaatgttaataattgagatttgcagaaaagaggaggaggtgggtttgg GTGCATTTCAAGGTGTTTTTTGCCAACACGGGTGCTTTGAagaaacaagcgtgagagacttgagtcag aagaacactttgtttttttttcttttctttttctaatcctattatatatatcaaaagactatgcttgt gagagtatgtatttatgaatgcgtaggaaaaaaaaagatgaatgctaaaagatagtagaccaatgtag cttttgcgaaaagcttagccatattatattaactaaactataaatgagacttggttttgcaaattatg NOI10 At5g48657: ttttttcttttgggtgtggttttagtcttctttttattttgattttctaaagcttattatcctacaag tgtagtatttattatacccacaatctctgagttttattccgtgagaatggagagtaattgtcaaatat ttgtagttctaaatatggatcctatatcatcctgtttaggacatatagtgttttgttttagaatcggc ggtatcatgagattttcttttgtttgacatttatgcgtaggaagatcacctaattctttttggtcatc
6 ttctgaacatcaatataattcatatgtaatgaaattatacttagattctacagatgaggtttctgttc aatcaaaaccaaaaaaaaagagataaggtttatgtttcttgcaccttaagattcagctctaaacattt agccaaaaacaaagtttcaactgtaaatctctggcatcaactccaaaataccgtatatatgagatttc gattttctaatatttatgtctcctactttcaatttaaatgcaagcctatgtttctcaaaatctttttt ttttggttcttgtggttatagaatcgtccacacgtgcccaaattcggagactggaccgaagatgctcc attcacggtcgtgttcgaaaaagcaagcaagagcaagaaaaatatgaacgtggccaacccgaatgaat ATCCAGATATGAATCCAAACGCTGCACAAAATCGAAATATGTCTAGGCCTGACCAACAACCACCAAAC CATAATGTTAGACCAAGACATGAAAGATTCAATAGCAGAGACGAAACCGAATTCAGACCGTCTCCTGC ACACAATGAAAGAAACAACAGAGTTAGATCAGTTCCTCCCACACCAGAAACATACAACCATCAAACAT ATGGTGGAGGTGGCAGATCA Insertion (SALK_080643C) ATGGGAAATCCAACAGAAATAAATAGACGGCAATCACGTGATCATGTCCCGGCGCGGCCAATACGCAA TCTTAGAGGGCAAAGTAGTGAAAGGgtatgcataactttaccattttatagtacaaaatatgtgtcac aactcataaatatatatcgctaagtaagtgtatttttctcaattaggtggcgactattcctccatttc CTGGAACGGGTTCTAACATGGAAAATCAGAGTTACACACTTATCTTTGACAAAGTGAAAGAAGATAGG AACCAAGCGAGATCTTACAATGGAACCGATCATAGCACCCCGACTCGACCCATCATCGACCAACATCA TCAACCATTACCTTCTAGTCCCAAGGTAGATAACCAAAATATCATTTCCCTTTCTTTCATAAATCCTC TTTTTTTACAATTGACGTTGTTATGTGTAGGGTTGCTGTTTTCCTCCATGGAGCCGAAAGGGAAGCTA ATGAAGATGATCATTTCCCTTTTTTACATAATCCTCTTTTTTTTTATTTGCTTCTGCTTGATTTTTTT TAATAATATCTTTGTTATTCTTATGGAGATGTAAggagtagtttttgctaaaacattatgcaacttct gttactctatttgcgtttttttgaacaatactcagattcatttataggggtgaatccttttattcacc tcctattataataaac NOI11 At3g07195: agagaagaaacggagacattcaaac Insertion (SALK_028339) tttacagagagtaacacgaatcgtcaacgcctttatatattacacacacattttatgtgtatctctgt gtttcagaatcagacctaagccattgttgaagcctttcctcatttctccatttagatcaagaaatatg GCAgtaagttcgaaaactcttttctatctaacaaaattgtgttatatccccctctcgtttgttatcat cttaaatcttgttatcaaaccatgttttatgacgaaaccatcagagtgttggaattttagcttgttga tccgaatttgtctgttcatttcccaatgattgtttaaattgtttgtttgatcatattagaacagagat ctgagtctttgtttcatatcagttgactttcgtatatattcttcttctgaattcaatctgttccagta tgtgttgagaagctaaagatagttctcataggtctgattagggttcatatacatatcgctttcttgca tctcaagtttgatattgaagtcattaaagctcaaatcacttttgaccattttctcaaaaacatttttt attgttgaatgattgattgcagaaccgtccacacgttccgaagtttggggactggaacaaccaagacc AACCATTCACGGTCGTCTTTGACAACGCAAGGACGAACAAACGACAAGACTTGTATGAATCTATAGAG AAACCAGAGACCAAACCTCAAGAGCTAGCTCCTCCTCCTCCTCAGCCTGCTCGTAGAATTCAAAAACC GGAAGCACCAAAACCGGTTAAGCAAGACACGCCGAGAGCACCTCCACCAACCGAAAAAAACAGAGTCA AAGCTCCACCGGCAGATCAACTCTACGGAGGCGGGGGCAGTGGAGGCGGAGGCAGGAGTGGAAGTGGA AGCGCAGGTGGTCTTTATGGAGGTTATGGAGGCGGATCAGTCGGGAACCAACGGCAGCCACCAGCTCC TCGTCCTACGCAGCCTAGACAAAATCTCAGAGGCGGCAACAACGGAAGGgtatgaaagacacaatata taaatgaacttggatctttgtttgcgatttgcattaacatgttattatgtgattgaagagtcacatta taatctaagatatatatatgatatgaatgaaaaagtttgcttatagtcattatatgatctgtagggag GGACAACGATACCGCCATTCCCAGGATCAGTTGGTGCAGGGGGGGACATGAGTTACACACAAATTTTC GAGAAAGTCAAAGAAGAGAGAAACGAAGGTGTCATCAGACCCAACGGAGGAACAGCCGGCAACACTCC TTCCCGCCCCATCAACAGTCAACATGATCAATCTACCAACCAAACCTCTTCCAAGgtagtttatttat taatatacaaatgtttaattacaaacacaactactttgtccatcgttagaatttgtgttttgatgtga attgggatgtgcagggctgctgcttctcatggtgccgcaggggaagtaaatattgaagacaatgtaca tatttatccctatttcgatatatctttttcttttgtaacaaaaaggagtgaagtttttgctgacaacg ttggagcttctacaatttttttttaaattattttaatttgtttctcaagtatggatggtgtagtttat ttgtgttgtgtatttgaattttgtaatgaaaaattgtttatttctgctaccgacaaaac NOI13 At4g35655: 6
7 aagcaagattttgaactctttttgcatcaaaattaggtaaaaacactaatcaaattacttgttaaaaa agg Insertion (SAIL_147_D02) cataataaatgacttgaagaaaatgtgatacaaatttagatttgttgaactcattgaccaggaggata ttatataaggtctctgattactaaacaagcatgtagagaaccaaagataacagtagcaccaaataaga aaacagagcagtcatggataagaagcattttgctataattagggttttacagggtatgacgcgcaaag accaaagattttgaacctcagaatgtttaagaagaaatgcgtaaagagaaatatcttgtctccgtgtt tcatacgagtacattccgtaggaacatttcataacaaaacttctctgtttctgctttattgaaatctt ccaactataaaatacaccctgagtccatcatcttttgatcaattagaacgatctcagaatagaagtca ttgaaaaagaagaacaacaataaaaggaagaaaataatttgtttttgagaaattttgctctgatttgt tgatcctgaagattgaaatggcatcgaatagtgacgcaaggcctttgccaaaattcggagaatgggat GTGAATGATCCGGCGACGGCAGAGGGATTCACCGTTATATTTAGTAAAGCCGGCGAGGATAAAAAAAC AGGTCGGAGCTCTAGCAAGGCACCTTCACAGAGAAAACAAGACGGCGTTAAACCAACCAAGAAATGGC TCTGTTTTACTTTTTCTTGAtttcctgtaaatttcttaaaacgtttgagaatggcattgagaaactct taccttagaaccttttctcatttgcttagcatttctctttataagatgttgcagagacgagaaagtag atagtttcttgtatatcttgtttgatgtttaatgtttaatgttatgtttcgtttgaagcttaagacta NOI14 At5g19473: aaacaatttgtactataaaggaaaaagtggaattttacaagagagatcgttctcctcctcgtcatgaa caccttcatcaatgtttaagaacatctttcttctaaaccaagcaatatctatttatggataaaagaag GGAGgcatgctctctaatttttttataaaatcttgtttcttttcttttatccaactattactaaatca tcgatctttatgatgtctgagttacaatcgagacatgttgagctttcatcggttaatgcgcgttttat agaatggggaaaattaaccgatgcaatctcaacatcgatctcattgttcttttttttttttccggaaa tatttggttccctaagcaaattatatgtttatctaacgagttttattcttggatacagaatgctgcat GGACTCCAGTTCCACAGTTCGGAGGGTGGGACCAAAAAGGACCAAATGATGCTACAAACTACTCGGTT GTCTTTTCTAAAGCTCGCGCAAACAGGAAGCAGAACAAAGCTGGTGTAAGACATTCTAGTTTGGGAAG CGAACAAGAACTCATGGTAAGTGTTCGTCGCAACCACCAACAACTTCACCATCGCCATGAAACTCAAG ACGATGACCCTGTCATGgtaattaaattatctctctctacttactcttccatggattctttggttcat attcaccttgtgatgatccttttccccttagtcaacgcgcatggatcatcacaaggtggtataacaat tcagga Insertion (SALK_110602) attgattttctctttgcgtcttttgtttgttttgtgtttttgcatgcgcctaaaaagaatatttcatt ccatttctttcttaactcagagacataattgaaccatgaatgttattcaacttaattatgagttaata catgcatcatagtatcaggtctcatttatgaatatgtaatgatagtgtatctttaaatttgatgttat agtgtattggcttgcagtttgtttccatggttttgaaatgatgtgttttgttcgaacaatgaatttat agttcgtttccattttcccattttctactttctttttatttgttaggaaaactttgtttttttttttg cattttttccataacatttcaataagcaaccatattttaacattatattcccacctaagctaaatttc attacttctattgttagataaaaatataattaaaccaactaaaagtctgaatcatattataatctttg cattatgttttaactatatgttgggagttacataaatttgttttgcttggagcctccaaactttgaag aggcaaacatgaacgaaatgtacactgcttttctagagcgaatatgaaaactccatctctagtttgta taacagtcgaaatcaccttgttatacgaagcattatgcaatatgtgaaatgtattaagaggaagcatg ttgtcaccgtcataagtctctctaacattactaatttaattaccatgcagaagaagaagaggatcttg ACCTACATTAATTGTTGTATAAGGCCAAACTAGggctttgttcttctacaaagtcggaatctttctct cttcagcgatatagcaaatatgtctcaagtgtgtaaagacagtagtgtaacgtttctgtaatcttctt tatcacacacaaaaaaaaaaaacgttctgttctatgtgcatcatctattaagatacccctggttgtgt tgtgttgtgatacattttgacttcttcctacatc Figure S2. Positions of T-DNA insertions in the NOI-genes. (A) Schematic representation of the T-DNA insertions with respect to the exon/intron genomic structures of the NOI genes. For exact location, see sequence information in (B). (generated with Exon-Intron Graphic Maker v4 ( (B) Sequencing of genomic regions flanking T-DNA insertions in NOI-genes. Genomic regions covering the NOI genes were exported from the TAIR SeqViewer tool ( T-DNA insertions are indicated, based on results obtained by sequencing PCR products of the respective regions. PCR reactions were performed using 7
8 the primers LBa1 (SALK lines) or LB3 (SAIL lines) plus respective RP primer (Table S2). PCR products were separated by gel electrophoresis and isolated bands were sequenced with the primers LBb1.3 (SALK lines) or LB2 (SAIL lines). Figure S3. Figure S3. Verification of knock out / knock down of transcript accumulation in noi-t-dna insertion lines. Leaf material of Col-0 and noi T-DNA insertion lines was used for quantitative RT-PCR experiments with NOI gene-specific primers (Table S2). 8
9 Figure S4. A CLUSTAL O(1.2.1) multiple sequence alignment of AvrRpt2 homologs Pst MKIAPVA Ea MKVSH Ac MRISSFSR Rs MKISHNGILGTHIPDT Mh 1 MKSRTYFSPNHSADR-FATRPNDDETATWVARLSGGQMQANDAHQQLSEPSLGGFDTDLD Sm 1 MDE-----TGVALRPYDASPENDHEVATWVARLSGGQMQANDAHQQLSEPSLGGFDTDLD Cf Bp Aa Pst 8 ---INHSPLSREVPSHAAPT----QAKQTNLQSEAGDLDARK-----SSASSP---ETR- Ea LTSPAPVVIEHQPRQSEKVSRDGDVI Ac 9 --AAGEN----EIPHTEEHQKKEGRTSQKMVSSEIFDQISKRDHTGVSSVAPRVNIGVAN Rs 17 SSQTGEA----SRHQEAQPNS-SGRPRPAGLA----GLQPKR-----NGLASRISVNKD- Mh 60 SDTIGRASMGESMHMPAQPSLGDGEPSSACLTPEAGVSQPPR-----LNVISRHS----- Sm 56 SGTIGRASMGESMHMPAQPSLGYGEFS Cf Bp Aa Pst 52 -ALLATKTVLGRHKIEVPAFGGWFKKKSSKHETGGSSANADSSSVASDSTEKPLFRLT-- Ea K-PWSQLPAAAPSFGGCFGKSKKSRG YDSGSSSGSRSNAGFRLN-- Ac 63 FDRNASKMSANVEVGNLPAFGGFFKKKSTG QSSSSGRGEESSTLPMFRLR-- Rs 62 ALPKRTDQTTQSAITALPAFGGWFGKKSSK-----SKGASSSGADASASGELPMFRLR-- Mh SAGQPGVISPRSSAPSFGGCFGSKAASSSHNEAKAASSSHNEASEATELPQFRVY-- Sm SAPPFGGWFSKKSS-----GSKAASSSHNEASEANELPQFRLY-- Cf Bp MVDVRLNVG Aa MGG GHLQSAGSCSSSG Pst Ea Ac Rs Mh Sm Cf Bp Aa Pst Ea Ac Rs Mh Sm Cf Bp Aa Pst Ea Ac Rs Mh Sm Cf Bp Aa HV--PYVSQGNERMGCWYACARMVGHSVEAGP--RLGLPELYEGREGPAGLQDFS HV--PYVSQQNERMGCWYACTRMLGHSISSGP--RLGLPELYDS-SGPQGLQQRE HV--PYVSQEDDRTGCWYACCRMLGYSVASGP--RLGLPELFDPNHGHSHLQSFE HV--PYVAQGSERTGCWYACARMLGHSVEAGP--RLGVPELFNPESGHSHLQHLH HV--PYISQGSERMGCWYACARMLASSVEAGP--RLGLPELYSPQSGHDGLQDPT HV--PYISQGSERSGCWYACARMVACSVETGP--RLGLPELYNPQSGHDGLQDLT MGCWYACARMIGHSAEAGP--RLGLPQRYSSDAGHQALKEPA 10 LVDQSGVKMTKSGSKEVKNACWYAAACMVSYYFRPGP--RRGIPKLWQADRGLPEA GSKLAVPYIPQN-GTRGCWEATMNMLRAYFGQPPLDKRQLAYLYDAEGVPKELNGQL Y191 D DVERFIHNEGLTRVDLPDNERFTHEELGALLYKHGPIIFGWKTPND--SWHMSVLTGVDK 125 DVLRLMRNENLAEVSLPESRQFSANELGNLLCRHGPIMFGWQTPAG--SWHMSVLTGIDK 164 HVERLLANEGLERVPLPSSQQFSLSELGALLYRHGPIIFGWQVPEG--YWHMSVMTGVDN 166 DVEQFIANEGLSKVPLPSSREFSHEQLGALLYRHGPIMFGWQTPQG--NWHMSVLTGVDH 216 HVEQFIVNEGLSKVDLPDSQEFSHEQLGDLLYRHGPIIFGWRTPEG--NSHMSVLTGVDK 172 HAEQFILNEGLSKLDLPDSQEFSHEELGELLYRHGPIIFGWQTPQG--NWHMSVLTGVDK 41 DIEQFISNEGLSRVNLPDSQAFSLEELVSLLEQHGPILFAWHPNQN--SGHMSVLIGVDK 64 RILELCAAEGLVPLDQPD--KITPEWLAAVLTDSGPIWAAGHFLDE--PGHVIVITGIQH 73 DYATRNRKAKLKNVPVPAGKSWDADKIKALLDQHGPLEARIESPHDELVAHAIVLTGIDE 218 ETSSITFHDPRQGPDLAMPLDYFNQRLAWQVPHAMLYR PNDAIIFHDPQRGPDLTMPLDSFNQRLAWRVPHAMLYSEN HTGSIIFHDPKKGPDITMPLAYFNQRLAWQVPHAMLYGGSS HTGHVVFHDPQQGPDLTMPLSYFNQRLAWQVPHAMLHR HTGRVVFHDPQKGPDLTMPLDYFNQRLAWQVPYAMLYRGVGAAP 230 HTGSVVFHDPQEGPDLTMPLDYFNQRLAWQVPHAMLYR KTSSVVYHDPQKGPDRGMPLSHFNQHLAWEVPYAMMHR SVVHYNDPEGPPARTMLLGTLDSG-RWKPD-GLLVKDRSRY- 133 D-GDVICHDPELGPRQKVSIEMLNAAFDWTSHRTALTAYNG--- 9
10 B Percent Identity Matrix of AvrRpt2 homologs created by CLUSTAL O(1.2.1) Aa Bp Ea Ac Pst Rs Cf Mh Sm 1: Aa : Bp : Ea : Ac : Pst : Rs : Cf : Mh : Sm Figure S4. Multiple sequence alignment and identity matrix of AvrRpt2 homologs. A,B. Multiple sequence alignments and identity matrix were done with CLUSTAL O(1.2.1) ( Sievers et al., 2011). A. Boxshade representation of the alignment ([LVI]PxFGxW= consensus AvrRpt2 cleavage site based on RIN4/NOIs; C/H/D= catalytic triad; = Y191 and D216 [reported to be important for virulence of Pst AvrRpt2]). B. Percent Identity Matrix of AvrRpt2 homologs (Aa= Acidovorax avenae subsp. avenae; Ac= Acidovorax citrulli; Bp= Burkholderia pyrrocinia; Cf= Collimonas fungivorans; Ea= Erwinia amylovora; Mh= Mesorhizobium huakuii; Pst= Pseudomonas syringae pv. tomato; Rs= Ralstonia solanacearum; Sm= Sinorhizobium medicae). Figure S5. Figure S5. Detection of low expression of the Bp AvrRpt2 homolog (longer exposure of Fig. 6C). See figure legend to Fig. 6C (*= weak expression of the Bp homolog; Bp= Burkholderia pyrrocinia; Cf= Collimonas fungivorans; Ea= Erwinia amylovora). 10
11 Figure S6. Figure S6. Western blot corresponding to experiment in Fig. 9A showing MAPK activation (αptepy), accumulation of MEKK1-YFP and HA-tagged CFP or AvrRpt2 variants. Note the three phosphorylated MAPK bands recognized by the α-ptepy antibodies are assumed to be MPK6, MPK3 and MPK4/MPK11 on the basis of the identical sizes to the flg22- activated MAPKs. 11
12 Table S1. AvrRpt2 homologs used in the study. organism Pseudomonas syringae pv. tomato JL1065 Mesorhizobium huakuii 7653R Erwinia amylovora ATCC Acidovorax avenae subsp. avenae ATCC Burkholderia pyrrocinia Lyc2 Collimonas fungivorans Acidovorax citrulli tw6 Sinorhizobium medicae WSM1369 Ralstonia solanacearum CMR15 GenBank-ID / NCBI 1 Reference Sequence CAA AID CBJ ADX KFL WP_ NZ_JXDJ NZ_AQUS NC_ (REGION: ) 1 The National Center for Biotechnology Information ( Table S2. List of primers and qpcr probes used in the study. name sequence purpose AvrRpt2_5 caccatgaaaattgctccagttgcc cloning of AvrRpt2 AvrRpt2_3 gcggtagagcattgcgtgtgg cloning of AvrRpt2 / qpcr (SYBRgreen) AvrRpt2_qPCR_5 cttttcacgatccccgacaggg qpcr (SYBRgreen) ACD11-like_5 gtacatcgccaaggttaagga qpcr (probe-based) ACD11-like_3 tctaggtcaagaatgttgtgtaacg qpcr (probe-based) ACD11-like-probe Universal ProbeLibrary probe #31 (Roche; cat. no ) qpcr (probe-based) BAP1_5 tcgtcaatctttcgattttgg qpcr (SYBRgreen) BAP1_3 gcctccacaaaccagtaacc qpcr (SYBRgreen) CAD5_5 gggagttatcaacaatccattaca qpcr (probe-based) CAD5_3 tgaagctccccgttatcact qpcr (probe-based) CAD5-probe Universal ProbeLibrary probe #82 (Roche; cat. no ) qpcr (probe-based) MPK3_5 tcaagcttcttcgtcatcttga qpcr (probe-based) MPK3_3 tgatttgatgaagatcagtatccatta qpcr (probe-based) MPK3-probe Universal ProbeLibrary probe #31 (Roche; cat. no ) qpcr (probe-based) MPK4_5 ggcgcttatggaattgtctg qpcr (probe-based) MPK4_3 ttgtcaaaagcattaccaatcttc qpcr (probe-based) MPK4-probe Universal ProbeLibrary probe #55 (Roche; cat. no ) qpcr (probe-based) MPK6_5 ataagcgatgagccagagtgta qpcr (probe-based) MPK6_3 cgcggtagattagttccttca qpcr (probe-based) MPK6-probe Universal ProbeLibrary probe #82 (Roche; cat. no ) qpcr (probe-based) MPK11_5 gcttctggcatcgtctgtg qpcr (probe-based) MPK11_3 ctcaaagttctcttagcgtcgat qpcr (probe-based) MPK11-probe FAM-tggaattcggagacgggtgagg-BHQ1 qpcr (probe-based) NHL10_5 acgccggacagtctagga qpcr (probe-based) NHL10_3 ccctaagcctgaacttgatctc qpcr (probe-based) 12
13 NHL10-probe Universal ProbeLibrary probe #55 (Roche; cat. no ) qpcr (probe-based) PCS1_5 acatatcaccgaggtgtatttaagc qpcr (probe-based) PCS1_3 gccaccaataggtgaaaagtg qpcr (probe-based) PCS1-probe Universal ProbeLibrary probe #95 (Roche; cat. no ) qpcr (probe-based) WRKY33_5 gggaaacccaaatccaaga qpcr (probe-based) WRKY33_3 gtttcccttcgtaggttgtga qpcr (probe-based) WRKY33-probe FAM-tgtggagagagcatcacacgacatg-BHQ1 qpcr (probe-based) WRKY53_5 cggaagtccgagaagtgaag qpcr (probe-based) WRKY53_3 tctgaccactttggtaacatcttt qpcr (probe-based) WRKY53-probe Universal ProbeLibrary probe #25 (Roche; cat. no ) qpcr (probe-based) ZAT12_5 ctttgggaggacacatgagg qpcr (probe-based) ZAT12_3 caaagcgcgtgtaaccaac qpcr (probe-based) ZAT12-probe Universal ProbeLibrary probe #66 (Roche; cat. no ) qpcr (probe-based) LBb1.3 ATTTTGCCGATTTCGGAAC sequencing of SALK T-DNA insertion lines LBa1 TGGTTCACGTAGTGGGCCATCG sequencing of SALK T-DNA insertion lines LB2 gcttcctattatatcttcccaaattaccaataca sequencing of SAIL T-DNA insertion lines LB3 tagcatctgaatttcataaccaatctcgatacac sequencing of SAIL T-DNA insertion lines NOI1_LP ttgttttgggattgaatgattg genotyping of SAIL_1173_C11 NOI1_RP ttgttttgatcggagttttgg genotyping of SAIL_1173_C11 NOI3_LP ggcttgagtattctggggaag genotyping of SALK_ NOI3_RP gaagataaaaagacgggacgg genotyping of SALK_ NOI4_LP atcgttttgatcatcgcaaac genotyping of SALK_042784C NOI4_RP ttttcggtttatccttgcctc genotyping of SALK_042784C NOI5_LP gtgacggttggaattgacaac genotyping of SALK_ NOI5_RP ctaattccaacaatggcaacg genotyping of SALK_ NOI6_LP aacaacaaccgcaatgagttc genotyping of SALK_024766C NOI6_RP attttaattgtgatgcagccg genotyping of SALK_024766C NOi7_LP ttttcattcggatcactgtcc genotyping of SALK_007305C NOI7_RP agctaatcaagctccaagcac genotyping of SALK_007305C NOI8_LP tcaaggtgacctagtagggattg genotyping of SALK_ NOI8_RP aatcaaacaaacaacgcgttc genotyping of SALK_ NOI9_LP caaaatctaaagccaacaagaaaac genotyping of SALK_150777C NOI9_RP aacccacctcctcctcttttc genotyping of SALK_150777C NOI10_LP ggtgaataaaaggattcacccc genotyping of SALK_080643C NOI10_RP tggcatcaactccaaaatacc genotyping of SALK_080643C NOI11a_LP ccagctaaccttagagacgcc genotyping of SALK_ NOI11a_RP aacttgagatgcaagaaagcg genotyping of SALK_ NOI13_LP tggtttgtttcaggtgacacc genotyping of SAIL_147_D02 NOI13_RP caaaagatgatggactcaggg genotyping of SAIL_147_D02 NOI14_LP agaacaaagccctagtttggc genotyping of SALK_ NOI14_RP tttccggaaatatttggttcc genotyping of SALK_ NOI1_qPCR_5 CAGAGAAACCAAAACTCCGAT qpcr (SYBRgreen) NOI1_qPCR_3 TTAACGGAAGCAAAACCATTTG qpcr (SYBRgreen) NOI3_qPCR_5 GGACGGAGTTCGACGAA qpcr (SYBRgreen) NOI3_qPCR_3 TTAAGCAAAAGTGAAACAGAGCCA qpcr (SYBRgreen) NOI4_qPCR_5 GACAGACACAAAGATGGATTTG qpcr (SYBRgreen) NOI4_qPCR_3 TCAAGAAAGACGACAATAAAACATAAA qpcr (SYBRgreen) NOI5_qPCR_5 GAACCTCCTAAAAACAACAACAA qpcr (SYBRgreen) NOI5_qPCR_3 TTAACGAAAACAAAGCCATTTCTTC qpcr (SYBRgreen) NOI6_qPCR_5 CAATGAGTTCTCTCACCATC qpcr (SYBRgreen) 13
14 NOI6_qPCR_3 TTAAGCTTTAACGCAACAGTTGAA qpcr (SYBRgreen) NOI7_qPCR_5 CATCATCAACCACATTCTCCT qpcr (SYBRgreen) NOI7_qPCR_3 TCAAGCTTTAACGCAGCAGTT qpcr (SYBRgreen) NOI8_qPCR_5 ATGAAGATCTCTACAAGATTCCTC qpcr (SYBRgreen) NOI8_qPCR_3 TCATGCGCATGGAACCAAAC qpcr (SYBRgreen) NOI9_qPCR_5 CCCACAACTTCTCTCAGTCAA qpcr (SYBRgreen) NOI9_qPCR_3 TCAAAGCACCCGTGTTGGC qpcr (SYBRgreen) NOI10_qPCR_5 GCTAATGAAGATGATCATTTCCCT qpcr (SYBRgreen) NOI10_qPCR_3 TTACATCTCCATAAGAATAACAAAGATATTAT qpcr (SYBRgreen) NOI11_qPCR_5 CTTCCCGCCCCATCAACA qpcr (SYBRgreen) NOI11_qPCR_3 TCAATATTTACTTCCCCTGCGG qpcr (SYBRgreen) NOI13_qPCR_5 GGTCGGAGCTCTAGCAAG qpcr (SYBRgreen) NOI13_qPCR_3 TCAAGAAAAAGTAAAACAGAGCCATTT qpcr (SYBRgreen) NOI14_qPCR_5 CACCATCGCCATGAAACTCAA qpcr (SYBRgreen) NOI14_qPCR_3 CTAGTTTGGCCTTATACAACAATTAA qpcr (SYBRgreen) RIN4_FL_5 atggcacgttcgaatgtacc RT-PCR RIN4 full length RIN4_FL_3 tcattttcctccaaagccaaag RT-PCR RIN4 full length RIN4-RCS1/2_5 atgaactgggaagctgaggag cloning and RT-PCR of RIN4 ACP2 RIN4-RCS1/2_3 tcaaccgaatttaggcaccac cloning and RT-PCR of RIN4 ACP2 RIN4-RCS2-end_5 atggactgggacgagaacaac cloning and RT-PCR of RIN4 ACP3 RIN4-RCS2-end_3 tcattttcctccaaagccaaag cloning and RT-PCR of RIN4 ACP3 18S_5 gatggtaggatagtggccta RT-PCR 18S rrna 18S_3 tggttgagactaggacgata RT-PCR 18S rrna AvrRpt2_Y191C_5 aaaaaagaagactgtgtaagcacgggccgattatatttgg site-directed mutagenesis of AvrRpt2 (Y191C) AvrRpt2_Y191C_3 aaaaaagaagaccttacacaacagtgcacccaactcttcgtgtg site-directed mutagenesis of AvrRpt2 (Y191C) AvrRpt2_D216E_5 aaaaaagaagactcgagaaagagacgtcgtccattacttttcac site-directed mutagenesis of AvrRpt2 (D216E) AvrRpt2_D216E_3 aaaaaagaagactttctcgacaccagtgaggaccgacat site-directed mutagenesis of AvrRpt2 (D216E) MKK1_T218D S224D_5 aaaaaaggtctcttgctaatgatttcgtgggcacata site-directed mutagenesis of MKK1 (T218D S224D) MKK1_T218D S224D_3 aaaaaaggtctctagcaagactactatcgcttgtca site-directed mutagenesis of MKK1 (T218D S224D) MKK2_T229E T235E_5 aaaaaaggtctcagcaaacgaatttgtggggact site-directed mutagenesis of MKK2 (T229E T235E) MKK2_T229E T235E_3 aaaaaaggtctctttgctaaacctgcttcgtttgtc site-directed mutagenesis of MKK2 (T229E T235E) StNOX fwd tcaatgcataggtatgaaggaatc qpcr (probe-based) StNOX rev tctcttcctagctagagcatcaaat qpcr (probe-based) StNOX probe FAM-tgaaggatggtctgaggttgaatcgcg-BHQ1 qpcr (probe-based) BcCutA-fwd ataagcgccgagcatgtg qpcr (SYBRgreen) BcCutA-rev gggatgacggaaaatagacg qpcr (SYBRgreen) Labeling: cacc= 5 -extension for directional cloning into pentr /D-TOPO vector (Thermo Scientific) GAAGAC/GGTCTC= type IIs restriction enzyme recognition site (mutagenesis was performed as described (Eschen-Lippold et al., 2014; Palm-Forster et al., 2012) FAM= 6-carboxyfluorescein BHQ1= black hole quencher 1 14
15 Table S3. List of vectors and DNA constructs used for transient expression in this study. DNA construct structure reference pugw14-[gw] p35s::[gw] C-term. HA-tag Nakagawa et al., 2007 pugw15-[gw] p35s::n-term. HA-tag [GW] Nakagawa et al., 2007 pugw18-[gw] p35s::n-term. c-myc-tag [GW] Nakagawa et al., 2007 pugw15-cfp p35s::ha-cfp this study pugw14-avrrpt2 p35s::avrrpt2-ha this study phbt-avrrpt2 H208A-HA p35s::avrrpt2 H208A-HA Cui et al., 2013 phbt-avrrpt2 C122A-FLAG p35s::avrrpt2 C122A-FLAG Cui et al., 2013 pugw14-avrrpt2 Y191C p35s::avrrpt2 Y191C-HA this study pugw14-avrrpt2 D216E p35s::avrrpt2 D216E-HA this study pugw18-rin4 p35s::c-myc-rin4 this study pugw15-rin4 F9A p35s::ha-rin4 F9A this study pugw15-rin4 F151A p35s::ha-rin4 F151A this study pugw15-rin4 F9A F151A p35s::ha-rin4 F9A F151A this study pugw15-rin4 ACP2 p35s::ha-rin4 ACP2 this study pugw15-rin4 ACP3 p35s::ha-rin4 ACP3 this study pugw18-noi1 p35s::c-myc-noi1 this study pugw18-noi2 p35s::c-myc-noi2 this study pugw18-noi3 p35s::c-myc-noi3 this study pugw18-noi4 p35s::c-myc-noi4 this study pugw18-noi5 p35s::c-myc-noi5 this study pugw18-noi6 p35s::c-myc-noi6 this study pugw18-noi7 p35s::c-myc-noi7 this study pugw18-noi8 p35s::c-myc-noi8 this study pugw18-noi9 p35s::c-myc-noi9 this study pugw18-noi10 p35s::c-myc-noi10 this study pugw18-noi11 p35s::c-myc-noi11 this study pugw18-noi12 p35s::c-myc-noi12 this study pugw18-noi13 p35s::c-myc-noi13 this study pugw18-noi14 p35s::c-myc-noi14 this study pugw14-aa avrrpt2 p35s::aa avrrpt2-ha this study pugw14-ac avrrpt2 p35s::ac avrrpt2-ha this study pugw14-bp avrrpt2 p35s::bp avrrpt2-ha this study pugw14-cf avrrpt2 p35s::cf avrrpt2-ha this study pugw14-ea avrrpt2 p35s::ea avrrpt2-ha this study pugw14-mh avrrpt2 p35s::mh avrrpt2-ha this study pugw14-rs avrrpt2 p35s::rs avrrpt2-ha this study pugw14-sm avrrpt2 p35s::sm avrrpt2-ha this study phbt-axr2-ha p35s::axr2-ha Cui et al., 2013 phbt-axr3-ha p35s::axr3-ha Cui et al., 2013 pexsg-yfp-mekk1 p35s::mekk1-yfp this study pugw14-mkk1 p35s::mkk1-ha this study pugw14-mkk1 T218D S224D p35s::mkk1 T218D S224D-HA this study pugw14-mkk2 p35s::mkk2-ha this study pugw14-mkk2 T229E T235E p35s::mkk2 T229E T235E-HA this study prt100myc-pcmkk5 p35s::c-myc-pcmkk5 Lee et al., 2004 Abbreviations: [GW]= Gateway Cloning cassette; Aa= Acidovorax avenae subsp. avenae ATCC 19860; Ac= Acidovorax citrulli strain tw6; Bp= Burkholderia pyrrocinia Lyc2; Cf= Collimonas fungivorans; Ea= Erwinia amylovora ATCC 49946; Mh= Mesorhizobium huakuii 7653R; Rs= Ralstonia solanacearum CMR15; Sm= Sinorhizobium medicae WSM1369; Pc= Petroselinum crispum 15
16 Supplemental Experimental Procedures Plant lines and treatment The plant lines used were rps2 (rps2-101c; Bent et al., 1994; Mindrinos et al., 1994), rps2rin4 (Mackey et al., 2003), rpm1rps2 (Mindrinos et al., 1994), rpm1rps2rin4 (Belkhadir et al., 2004), DEX-T7-RIN4 and DEX-T7-RIN4 CCC>AAA (dexamethasone-inducible expression of T7- tagged RIN4 and RIN4 C203A/C204A/C205A in Col-0 background, respectively; Afzal et al., 2011), DEX-avrRpt2 (dexamethasone-inducible expression of AvrRpt2 in rps2-101c background; Mackey et al., 2003), 35S::axr2-1/DEX::avrRpt2/rps2 (over-expression of axr2-1 and dexamethasone-inducible expression of AvrRpt2 in the rps2 knock out background ; lines #20 and #23; Cui et al., 2013). To identify noi-gene knock out plants, the following T-DNA insertion lines were obtained from Nottingham Arabidopsis Stock Centre (Sessions et al., 2002; Alonso et al., 2003): noi1 - At5g SAIL_1173_C11; noi4 - At5g SALK_042784C; noi5 - At3g SALK_118446; noi7 - At5g SALK_007305C; noi8 - At5g SALK_101426; noi9 - At5g SALK_150777C; noi10 - At5g SALK_080643C; noi11 - At3g SALK_028339; noi13 - At4g SAIL_147_D02; noi14 - At5g SALK_ DEX-avrRpt2 plants were sprayed with 20 µm dexamethasone to induce AvrRpt2 expression. Five to six hours later, leaves were infiltrated with 1 µm flg22 to induce MAPK activation. Protoplast work For Western-blot analyses, 300 µl protoplast samples were each treated with 100 nm flg22 (or an equal volume of water) and harvested by centrifugation at the indicated time points. Media were discarded and cell pellets were snap-frozen in liquid nitrogen. To induce expression of RIN4 and RIN4 CCC>AAA in protoplasts derived from the respective DEX-inducible lines, 16
17 protoplasts were incubated after transfection in the presence of 10 µm dexamethasone overnight. Protein work Protein extraction from plant material was performed as described (Lee et al., 2004). In case of protoplasts, standard SDS loading dye was directly added to the pelleted samples. The antibodies used for immunoblotting were: α-ptepy (phospho-p44/42 MAPK [ERK1/2] [Thr202/Tyr204]; Cell Signaling Technology), α-mpk4 (Anti-AtMPK4; Sigma), α-ha (α-ha.11; Eurogentec), α-flag (FLAG Epitope Tag [DYKDDDDK] Antibody [FG4R]; Thermo Scientific), or α-rin4 (Mackey et al., 2002). Quantitative realtime PCR and semi-quantitative RT-PCR Genomic DNA was isolated with the GenElute Plant Genomic DNA Miniprep Kit (Sigma). RNA was isolated with standard Trizol reagent, treated with DNAseI and cdna synthesis was performed with the RevertAid kit (Thermo Scientific). For quantitative realtime PCR, amplicons from either genomic DNA samples (in case of Botrytis cinerea biomass quantification) or cdna samples (for expression analyses) as templates were analyzed with an Mx3005P qpcr system (Agilent) using the Maxima Probe qpcr and Maxima SYBR Green qpcr Master Mixes (Thermo Scientific). For the Botrytis cinerea experiments, an external control (plasmid clone of a potato StNOX gene) was spiked into the collected leaf material prior to genomic DNA extraction. For fungal biomass quantification, primers for Botrytis cinerea Cutinase A (accession number Z69264) gene were used. BcCutA data were normalized to corresponding StNOX data. Relative gene expression values of the genes-of-interest were calculated with the comparative C T method (Schmittgen and Livak, 2008) using the reference gene PP2A (AT1G13320; Czechowski et al. 2005). Primers and hydrolysis probes are listed in Table S2. 17
18 Semi-quantitative RT-PCR was performed using DreamTaq DNA Polymerase (Thermo Scientific) with the primers listed in Table S2. Callose detection Callose staining was performed with aniline blue (Adam & Somerville; 1996). Photographs were taken using a Nikon multizoom AZ100 microscope (Nikon Instruments) with the following UV filter combination: nm excitation filter, 400 nm (LP) dichromatic mirror and 405 nm (LP) barrier filter. Reactive oxygen species measurement Production of reactive oxygen species was measured according to (Gomez-Gomez et al., 1999) using 3 mm leaf discs in a 96-well plate format measured in 2 min intervals (60 min total time) with a Luminoskan Ascent 2.1 device (Thermo Scientific). Phylogenetic analysis Phylogenetic analysis was performed with MEGA4 software (Tamura et al., 2007) using the Neighbor-Joining method (Saitou and Nei, 1987). The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches (Felsenstein, 1985). The trees are drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method (Zuckerkandl and Pauling, 1965) and are in the units of the number of amino acid substitutions per site. The optimal trees with the sum of branch lengths = (Fig. 3A) and (Fig. 6A) are shown. All positions containing gaps and missing data were eliminated from the dataset 18
19 (complete deletion option). There were a total of 53 (Fig. 3A) and 126 (Fig. 6A) positions in the final dataset. Supplemental References Adam, L., and Somerville, S.C. (1996). Genetic characterization of five powdery mildew disease resistance loci in Arabidopsis thaliana. Plant J 9, Afzal, A.J., da Cunha, L., and Mackey, D. (2011). Separable fragments and membrane tethering of Arabidopsis RIN4 regulate its suppression of PAMP-triggered immunity. The Plant Cell 23, Alonso, J.M., Stepanova, A.N., Leisse, T.J., Kim, C.J., Chen, H., Shinn, P., Stevenson, D.K., Zimmerman, J., Barajas, P., Cheuk, R., Gadrinab, C., Heller, C., Jeske, A., Koesema, E., Meyers, C.C., Parker, H., Prednis, L., Ansari, Y., Choy, N., Deen, H., Geralt, M., Hazari, N., Hom, E., Karnes, M., Mulholland, C., Ndubaku, R., Schmidt, I., Guzman, P., Aguilar-Henonin, L., Schmid, M., Weigel, D., Carter, D.E., Marchand, T., Risseeuw, E., Brogden, D., Zeko, A., Crosby, W.L., Berry, C.C., and Ecker, J.R. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, Belkhadir, Y., Nimchuk, Z., Hubert, D.A., Mackey, D., and Dangl, J.L. (2004). Arabidopsis RIN4 negatively regulates disease resistance mediated by RPS2 and RPM1 downstream or independent of the NDR1 signal modulator and is not required for the virulence functions of bacterial type III effectors AvrRpt2 or AvrRpm1. The Plant Cell 16, Bent, A.F., Kunkel, B.N., Dahlbeck, D., Brown, K.L., Schmidt, R., Giraudat, J., Leung, J., and Staskawicz, B.J. (1994). RPS2 of Arabidopsis thaliana: a leucine-rich repeat class of plant disease resistance genes. Science 265, Czechowski, T., Stitt, M., Altmann, T., Udvardi, M.K., and Scheible, W.R. (2005). Genomewide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139, Chen, Z., Agnew, J.L., Cohen, J.D., He, P., Shan, L., Sheen, J., and Kunkel, B.N. (2007). Pseudomonas syringae type III effector AvrRpt2 alters Arabidopsis thaliana auxin physiology. Proc Natl Acad Sci U S A 104, Cui, F., Wu, S., Sun, W., Coaker, G., Kunkel, B., He, P., and Shan, L. (2013). The Pseudomonas syringae type III effector AvrRpt2 promotes pathogen virulence via stimulating Arabidopsis auxin/indole acetic acid protein turnover. Plant physiology 162, Eschen-Lippold, L., Bauer, N., Löhr, J., Palm-Forster, M.A.T., and Lee, J. (2014). Rapid Mutagenesis-Based Analysis of Phosphorylation Sites in Mitogen-Activated Protein Kinase Substrates. In Plant MAP Kinases, G. Komis, and J. Samaj, eds. (Springer New York), pp Felsenstein, J. (1985). Confidence-Limits on Phylogenies - an Approach Using the Bootstrap. Evolution 39, Gómez-Gómez, L., Felix, G. and Boller, T. (1999). A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J 18, Lee, J., Rudd, J.J., Macioszek, V.K., and Scheel, D. (2004). Dynamic changes in the localization of MAPK cascade components controlling pathogenesis-related (PR) gene expression during innate immunity in parsley. J Biol Chem 279,
20 Mackey, D., Holt, B.F., Wiig, A., and Dangl, J.L. (2002). RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108, Mackey, D., Belkhadir, Y., Alonso, J.M., Ecker, J.R., and Dangl, J.L. (2003). Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112, Mindrinos, M., Katagiri, F., Yu, G.L., and Ausubel, F.M. (1994). The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide-binding site and leucinerich repeats. Cell 78, Nakagawa, T., Kurose, T., Hino, T., Tanaka, K., Kawamukai, M., Niwa, Y., Toyooka, K., Matsuoka, K., Jinbo, T., and Kimura, T. (2007). Development of series of gateway binary vectors, pgwbs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104, Palm-Forster, M.A.T., Eschen-Lippold, L., and Lee, J. (2012). A mutagenesis-based screen to rapidly identify phosphorylation sites in mitogen-activated protein kinase substrates. Anal Biochem 427, Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4, Schmittgen, T.D., and Livak, K.J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3, Sessions, A., Burke, E., Presting, G., Aux, G., McElver, J., Patton, D., Dietrich, B., Ho, P., Bacwaden, J., Ko, C., Clarke, J.D., Cotton, D., Bullis, D., Snell, J., Miguel, T., Hutchison, D., Kimmerly, B., Mitzel, T., Katagiri, F., Glazebrook, J., Law, M., and Goff, S.A. (2002). A highthroughput Arabidopsis reverse genetics system. The Plant Cell 14, Sievers, F., Wilm, A., Dineen, D., Gibson, T.J., Karplus, K., Li, W.Z., Lopez, R., McWilliam, H., Remmert, M., Soding, J., Thompson, J.D., and Higgins, D.G. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7. Tamura, K., Dudley, J., Nei, M., and Kumar, S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24, Zuckerkandl, E., and Pauling, L. (1965). Evolutionary divergence and convergence in proteins. In Evolving Genes and Proteins, V. Bryson and H.J. Vogel, eds (New York: Academic Press), pp
Experimental Identification and Computational Characterization of a Novel. Extracellular Metalloproteinase Produced by Clostridium sordellii
 Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 207 Supplementary Information Experimental Identification and Computational Characterization of
Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 207 Supplementary Information Experimental Identification and Computational Characterization of
Plasmid Name: pmm290. Aliases: none known. Length: bp. Constructed by: Mike Moser/Cristina Swanson. Last updated: 17 August 2009
 Plasmid Name: pmm290 Aliases: none known Length: 11707 bp Constructed by: Mike Moser/Cristina Swanson Last updated: 17 August 2009 Description and application: This is a mammalian expression vector for
Plasmid Name: pmm290 Aliases: none known Length: 11707 bp Constructed by: Mike Moser/Cristina Swanson Last updated: 17 August 2009 Description and application: This is a mammalian expression vector for
tgg agg Supplementary Figure S1.
 ttaggatattcggtgaggtgatatgtctctgtttggaaatgtctccgccattaactcaag tggaaagtgtatagtaatgaatctttcaagcacacagatcacttcaaaagactgtttcaa catcacctcaggacaaaaagatgtactctcatttggatgctgtgatgccatgggtcacag attgcaattcccaagtgcccgttcttttacaccaaaatcaaagaagaatatctccccttt
ttaggatattcggtgaggtgatatgtctctgtttggaaatgtctccgccattaactcaag tggaaagtgtatagtaatgaatctttcaagcacacagatcacttcaaaagactgtttcaa catcacctcaggacaaaaagatgtactctcatttggatgctgtgatgccatgggtcacag attgcaattcccaagtgcccgttcttttacaccaaaatcaaagaagaatatctccccttt
Supporting information
 Supporting information Figure S1. Carotenoid biosynthesis pathway in papaya fruit which is adopted from Blas et al. (2010) (reference 4) and Nisar et al. (2015) (reference 5). Carotenoids are synthesized
Supporting information Figure S1. Carotenoid biosynthesis pathway in papaya fruit which is adopted from Blas et al. (2010) (reference 4) and Nisar et al. (2015) (reference 5). Carotenoids are synthesized
Supporting Information for
 Supporting Information for Analysis of Sogatella furcifera proteome that interact with P10 protein of Southern rice black-streaked dwarf virus Win Than*, Faliang Qin*, Wenwen Liu, Xifeng Wang ** State
Supporting Information for Analysis of Sogatella furcifera proteome that interact with P10 protein of Southern rice black-streaked dwarf virus Win Than*, Faliang Qin*, Wenwen Liu, Xifeng Wang ** State
Methods S1. Sequences relevant to the constructed strains, Related to Figures 1-6.
 Methods S1. Sequences relevant to the constructed strains, Related to Figures 1-6. A. Promoter Sequences Gal4 binding sites are highlighted in the color referenced in Figure 1A when possible. Site 1: red,
Methods S1. Sequences relevant to the constructed strains, Related to Figures 1-6. A. Promoter Sequences Gal4 binding sites are highlighted in the color referenced in Figure 1A when possible. Site 1: red,
Supplementary information: Biocatalysis on the surface of Escherichia coli: melanin pigmentation of the cell. exterior
 Supplementary information: Biocatalysis on the surface of Escherichia coli: melanin pigmentation of the cell exterior Martin Gustavsson, David Hörnström, Susanna Lundh, Jaroslav Belotserkovsky, Gen Larsson
Supplementary information: Biocatalysis on the surface of Escherichia coli: melanin pigmentation of the cell exterior Martin Gustavsson, David Hörnström, Susanna Lundh, Jaroslav Belotserkovsky, Gen Larsson
FETAL FIBROBLASTS, PASSAGE 10
 Double-stranded methylation patterns of a 104-bp L1 promoter in DNAs from fetal fibroblast passages 10, 14, 17, and 22 using barcoded hairpinbisulfite PCR. Fifteen L1 sequences were analyzed for passages
Double-stranded methylation patterns of a 104-bp L1 promoter in DNAs from fetal fibroblast passages 10, 14, 17, and 22 using barcoded hairpinbisulfite PCR. Fifteen L1 sequences were analyzed for passages
Strain or plasmid Description a Reference or source b. 168 trpc2 Laboratory. 1A780 trpc2 sigb::spc BGSC
 TABLE S1 Bacterial strains and plasmids Strain or plasmid Description a Reference or source b B. subtilis 168 trpc2 Laboratory stock 1A780 trpc2 sigb::spc BGSC BM1010 trpc2 htpx::pgs1582; Em r BM1302 trpc2
TABLE S1 Bacterial strains and plasmids Strain or plasmid Description a Reference or source b B. subtilis 168 trpc2 Laboratory stock 1A780 trpc2 sigb::spc BGSC BM1010 trpc2 htpx::pgs1582; Em r BM1302 trpc2
Plasmid construction PCR reactions were carried out with Pfu or Pfu turbo polymerases (Stratagene) unless otherwise stated.
 Expanding the Genetic Code of Yeast for Incorporation of Diverse Unnatural Amino Acids via a Pyrrolysyl-tRNA Synthetase/tRNA Pair Susan M. Hancock 1, Rajendra Uprety 2, Alexander Deiters 2 & Jason W. Chin
Expanding the Genetic Code of Yeast for Incorporation of Diverse Unnatural Amino Acids via a Pyrrolysyl-tRNA Synthetase/tRNA Pair Susan M. Hancock 1, Rajendra Uprety 2, Alexander Deiters 2 & Jason W. Chin
MALE ADULT FIBROBLAST LINE (82-6hTERT)
 Double-stranded methylation patterns of a 104-bp L1 promoter in DNAs from male and female fibroblasts, male leukocytes and female lymphoblastoid cells using hairpin-bisulfite PCR. Fifteen L1 sequences
Double-stranded methylation patterns of a 104-bp L1 promoter in DNAs from male and female fibroblasts, male leukocytes and female lymphoblastoid cells using hairpin-bisulfite PCR. Fifteen L1 sequences
Functional Genomics & Proteomics
 Functional Genomics & Proteomics Genome Sequences TCACAATTTAGACATCTAGTCTTCCACTTAAGCATATTTAGATTGTTTCCAGTTTTCAGCTTTTATGACTAAATCTTCTAAAATTGTTTTTCCCTAAATGTATATTTTAATTTGTCTCAGGAGTAGAATTTCTGAGTCATAAAGCGGT CATATGTATAAATTTTAGGTGCCTCATAGCTCTTCAAATAGTCATCCCATTTTATACATCCAGGCAATATATGAGAGTTCTTGGTGCTCCACATCTTAGCTAGGATTTGATGTCAACCAGTCTCTTTAATTTAGATATTCTAGTACAT
Functional Genomics & Proteomics Genome Sequences TCACAATTTAGACATCTAGTCTTCCACTTAAGCATATTTAGATTGTTTCCAGTTTTCAGCTTTTATGACTAAATCTTCTAAAATTGTTTTTCCCTAAATGTATATTTTAATTTGTCTCAGGAGTAGAATTTCTGAGTCATAAAGCGGT CATATGTATAAATTTTAGGTGCCTCATAGCTCTTCAAATAGTCATCCCATTTTATACATCCAGGCAATATATGAGAGTTCTTGGTGCTCCACATCTTAGCTAGGATTTGATGTCAACCAGTCTCTTTAATTTAGATATTCTAGTACAT
State of the Union... Functional Genomics Research Stream. Molecular Biology. Genomics. Computational Biology
 Functional Genomics Research Stream State of the Union... Research Meeting: February 16, 2010 Functional Genomics & Research Report III Concepts Genomics Molecular Biology Computational Biology Genome
Functional Genomics Research Stream State of the Union... Research Meeting: February 16, 2010 Functional Genomics & Research Report III Concepts Genomics Molecular Biology Computational Biology Genome
Stormwater filtration unit
 Stormwater filtration unit Background, concept and applied design work Olli Hakala 2018 WSP Finland Aalto university Kyttä ym. 2014. Veden äärellä kysely, ENTJUSTESS-hanke. Aalto yliopisto. STORMWATER
Stormwater filtration unit Background, concept and applied design work Olli Hakala 2018 WSP Finland Aalto university Kyttä ym. 2014. Veden äärellä kysely, ENTJUSTESS-hanke. Aalto yliopisto. STORMWATER
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
Other approaches to restrict multipliers
 Other approaches to restrict multipliers Heikki Tikanmäki Optimointiopin seminaari 10.10.2007 Contents Short revision (6.2) Another Assurance Region Model (6.3) Cone-Ratio Method (6.4) An Application of
Other approaches to restrict multipliers Heikki Tikanmäki Optimointiopin seminaari 10.10.2007 Contents Short revision (6.2) Another Assurance Region Model (6.3) Cone-Ratio Method (6.4) An Application of
lpar1 IPB004065, IPB002277, and IPB Restriction Enyzme Differences from REBASE Gained in Variant Lost from Reference
 lpar1 IPB465, IPB2277, and IPB5385 Genomic Sequence Coding Sequence For help interpreting these results, view the PARSENP Introduction page. # View On Sequence Nucleotide Change Effect Restriction Enyzme
lpar1 IPB465, IPB2277, and IPB5385 Genomic Sequence Coding Sequence For help interpreting these results, view the PARSENP Introduction page. # View On Sequence Nucleotide Change Effect Restriction Enyzme
Basset: Learning the regulatory code of the accessible genome with deep convolutional neural networks. David R. Kelley
 Basset: Learning the regulatory code of the accessible genome with deep convolutional neural networks David R. Kelley DNA codes for complex life. How? Kundaje et al. Integrative analysis of 111 reference
Basset: Learning the regulatory code of the accessible genome with deep convolutional neural networks David R. Kelley DNA codes for complex life. How? Kundaje et al. Integrative analysis of 111 reference
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
16. Allocation Models
 16. Allocation Models Juha Saloheimo 17.1.27 S steemianalsin Optimointiopin seminaari - Sks 27 Content Introduction Overall Efficienc with common prices and costs Cost Efficienc S steemianalsin Revenue
16. Allocation Models Juha Saloheimo 17.1.27 S steemianalsin Optimointiopin seminaari - Sks 27 Content Introduction Overall Efficienc with common prices and costs Cost Efficienc S steemianalsin Revenue
Efficiency change over time
 Efficiency change over time Heikki Tikanmäki Optimointiopin seminaari 14.11.2007 Contents Introduction (11.1) Window analysis (11.2) Example, application, analysis Malmquist index (11.3) Dealing with panel
Efficiency change over time Heikki Tikanmäki Optimointiopin seminaari 14.11.2007 Contents Introduction (11.1) Window analysis (11.2) Example, application, analysis Malmquist index (11.3) Dealing with panel
Metsälamminkankaan tuulivoimapuiston osayleiskaava
 VAALAN KUNTA TUULISAIMAA OY Metsälamminkankaan tuulivoimapuiston osayleiskaava Liite 3. Varjostusmallinnus FCG SUUNNITTELU JA TEKNIIKKA OY 12.5.2015 P25370 SHADOW - Main Result Assumptions for shadow calculations
VAALAN KUNTA TUULISAIMAA OY Metsälamminkankaan tuulivoimapuiston osayleiskaava Liite 3. Varjostusmallinnus FCG SUUNNITTELU JA TEKNIIKKA OY 12.5.2015 P25370 SHADOW - Main Result Assumptions for shadow calculations
Returns to Scale II. S ysteemianalyysin. Laboratorio. Esitelmä 8 Timo Salminen. Teknillinen korkeakoulu
 Returns to Scale II Contents Most Productive Scale Size Further Considerations Relaxation of the Convexity Condition Useful Reminder Theorem 5.5 A DMU found to be efficient with a CCR model will also be
Returns to Scale II Contents Most Productive Scale Size Further Considerations Relaxation of the Convexity Condition Useful Reminder Theorem 5.5 A DMU found to be efficient with a CCR model will also be
rapid evolution in copy number, location and sequence, with diverse turnover mechanisms.
 Repetitive DNA and its evolution: genomic strategies to understand the processes and exploit biodiversity rapid evolution in copy number, location and sequence, with diverse turnover mechanisms. often
Repetitive DNA and its evolution: genomic strategies to understand the processes and exploit biodiversity rapid evolution in copy number, location and sequence, with diverse turnover mechanisms. often
Eukaryotic Comparative Genomics
 Eukaryotic Comparative Genomics Detecting Conserved Sequences Charles Darwin Motoo Kimura Evolution of Neutral DNA A A T C TA AT T G CT G T GA T T C A GA G T A G CA G T GA AT A GT C T T T GA T GT T G T
Eukaryotic Comparative Genomics Detecting Conserved Sequences Charles Darwin Motoo Kimura Evolution of Neutral DNA A A T C TA AT T G CT G T GA T T C A GA G T A G CA G T GA AT A GT C T T T GA T GT T G T
Tynnyrivaara, OX2 Tuulivoimahanke. ( Layout 9 x N131 x HH145. Rakennukset Asuinrakennus Lomarakennus 9 x N131 x HH145 Varjostus 1 h/a 8 h/a 20 h/a
 , Tuulivoimahanke Layout 9 x N131 x HH145 Rakennukset Asuinrakennus Lomarakennus 9 x N131 x HH145 Varjostus 1 h/a 8 h/a 20 h/a 0 0,5 1 1,5 km 2 SHADOW - Main Result Assumptions for shadow calculations
, Tuulivoimahanke Layout 9 x N131 x HH145 Rakennukset Asuinrakennus Lomarakennus 9 x N131 x HH145 Varjostus 1 h/a 8 h/a 20 h/a 0 0,5 1 1,5 km 2 SHADOW - Main Result Assumptions for shadow calculations
7.4 Variability management
 7.4 Variability management time... space software product-line should support variability in space (different products) support variability in time (maintenance, evolution) 1 Product variation Product
7.4 Variability management time... space software product-line should support variability in space (different products) support variability in time (maintenance, evolution) 1 Product variation Product
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.9.269
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.9.269
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
( ( OX2 Perkkiö. Rakennuskanta. Varjostus. 9 x N131 x HH145
 OX2 9 x N131 x HH145 Rakennuskanta Asuinrakennus Lomarakennus Liike- tai julkinen rakennus Teollinen rakennus Kirkko tai kirkollinen rak. Muu rakennus Allas Varjostus 1 h/a 8 h/a 20 h/a 0 0,5 1 1,5 2 km
OX2 9 x N131 x HH145 Rakennuskanta Asuinrakennus Lomarakennus Liike- tai julkinen rakennus Teollinen rakennus Kirkko tai kirkollinen rak. Muu rakennus Allas Varjostus 1 h/a 8 h/a 20 h/a 0 0,5 1 1,5 2 km
Capacity Utilization
 Capacity Utilization Tim Schöneberg 28th November Agenda Introduction Fixed and variable input ressources Technical capacity utilization Price based capacity utilization measure Long run and short run
Capacity Utilization Tim Schöneberg 28th November Agenda Introduction Fixed and variable input ressources Technical capacity utilization Price based capacity utilization measure Long run and short run
Alternative DEA Models
 Mat-2.4142 Alternative DEA Models 19.9.2007 Table of Contents Banker-Charnes-Cooper Model Additive Model Example Data Home assignment BCC Model (Banker-Charnes-Cooper) production frontiers spanned by convex
Mat-2.4142 Alternative DEA Models 19.9.2007 Table of Contents Banker-Charnes-Cooper Model Additive Model Example Data Home assignment BCC Model (Banker-Charnes-Cooper) production frontiers spanned by convex
Efficient regeneration system and Agrobacteriummediated transformation of Vetiver. PT 1
 Efficient regeneration system and Agrobacteriummediated transformation of Vetiver PT www.acolor.net 1 Outline: Part I: Why (Background) Part II: Some concrete work we have done Part III: Prospects to further
Efficient regeneration system and Agrobacteriummediated transformation of Vetiver PT www.acolor.net 1 Outline: Part I: Why (Background) Part II: Some concrete work we have done Part III: Prospects to further
TM ETRS-TM35FIN-ETRS89 WTG
 VE1 SHADOW - Main Result Calculation: 8 x Nordex N131 x HH145m Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please
VE1 SHADOW - Main Result Calculation: 8 x Nordex N131 x HH145m Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Calculation: N117 x 9 x HH141 Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG
SHADOW - Main Result Calculation: N117 x 9 x HH141 Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG
WindPRO version joulu 2012 Printed/Page :47 / 1. SHADOW - Main Result
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
WindPRO version joulu 2012 Printed/Page :42 / 1. SHADOW - Main Result
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table 13.6.2013 19:42 / 1 Minimum
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table 13.6.2013 19:42 / 1 Minimum
KONEISTUSKOKOONPANON TEKEMINEN NX10-YMPÄRISTÖSSÄ
 KONEISTUSKOKOONPANON TEKEMINEN NX10-YMPÄRISTÖSSÄ https://community.plm.automation.siemens.com/t5/tech-tips- Knowledge-Base-NX/How-to-simulate-any-G-code-file-in-NX- CAM/ta-p/3340 Koneistusympäristön määrittely
KONEISTUSKOKOONPANON TEKEMINEN NX10-YMPÄRISTÖSSÄ https://community.plm.automation.siemens.com/t5/tech-tips- Knowledge-Base-NX/How-to-simulate-any-G-code-file-in-NX- CAM/ta-p/3340 Koneistusympäristön määrittely
Supplementary Material : Baker et al. 1
 Supplementary Material : Baker et al. 1 Supplementary Material Title: Species identity and human consumption of beaked whales in the Gilbert Islands, Republic of Kiribati Authors: C. Scott Baker, Al Hutt,
Supplementary Material : Baker et al. 1 Supplementary Material Title: Species identity and human consumption of beaked whales in the Gilbert Islands, Republic of Kiribati Authors: C. Scott Baker, Al Hutt,
LX 70. Ominaisuuksien mittaustulokset 1-kerroksinen 2-kerroksinen. Fyysiset ominaisuudet, nimellisarvot. Kalvon ominaisuudet
 LX 70 % Läpäisy 36 32 % Absorptio 30 40 % Heijastus 34 28 % Läpäisy 72 65 % Heijastus ulkopuoli 9 16 % Heijastus sisäpuoli 9 13 Emissiivisyys.77.77 Auringonsuojakerroin.54.58 Auringonsäteilyn lämmönsiirtokerroin.47.50
LX 70 % Läpäisy 36 32 % Absorptio 30 40 % Heijastus 34 28 % Läpäisy 72 65 % Heijastus ulkopuoli 9 16 % Heijastus sisäpuoli 9 13 Emissiivisyys.77.77 Auringonsuojakerroin.54.58 Auringonsäteilyn lämmönsiirtokerroin.47.50
( ,5 1 1,5 2 km
 Tuulivoimala Rakennukset Asuinrakennus Liikerak. tai Julkinen rak. Lomarakennus Teollinen rakennus Kirkollinen rakennus Varjostus "real case" h/a 1 h/a 8 h/a 20 h/a 4 5 3 1 2 6 7 8 9 10 0 0,5 1 1,5 2 km
Tuulivoimala Rakennukset Asuinrakennus Liikerak. tai Julkinen rak. Lomarakennus Teollinen rakennus Kirkollinen rakennus Varjostus "real case" h/a 1 h/a 8 h/a 20 h/a 4 5 3 1 2 6 7 8 9 10 0 0,5 1 1,5 2 km
,0 Yes ,0 120, ,8
 SHADOW - Main Result Calculation: Alue 2 ( x 9 x HH120) TuuliSaimaa kaavaluonnos Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered
SHADOW - Main Result Calculation: Alue 2 ( x 9 x HH120) TuuliSaimaa kaavaluonnos Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered
Underpinning Phage Arbitrium Communication Systems
 Molecular Cell, Volume 74 Supplemental Information Deciphering the Molecular Mechanism Underpinning Phage Arbitrium Communication Systems Francisca Gallego del Sol, José R. Penadés, and Alberto Marina
Molecular Cell, Volume 74 Supplemental Information Deciphering the Molecular Mechanism Underpinning Phage Arbitrium Communication Systems Francisca Gallego del Sol, José R. Penadés, and Alberto Marina
1. SIT. The handler and dog stop with the dog sitting at heel. When the dog is sitting, the handler cues the dog to heel forward.
 START START SIT 1. SIT. The handler and dog stop with the dog sitting at heel. When the dog is sitting, the handler cues the dog to heel forward. This is a static exercise. SIT STAND 2. SIT STAND. The
START START SIT 1. SIT. The handler and dog stop with the dog sitting at heel. When the dog is sitting, the handler cues the dog to heel forward. This is a static exercise. SIT STAND 2. SIT STAND. The
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table 22.12.2014 11:33 / 1 Minimum
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table 22.12.2014 11:33 / 1 Minimum
Use of spatial data in the new production environment and in a data warehouse
 Use of spatial data in the new production environment and in a data warehouse Nordic Forum for Geostatistics 2007 Session 3, GI infrastructure and use of spatial database Statistics Finland, Population
Use of spatial data in the new production environment and in a data warehouse Nordic Forum for Geostatistics 2007 Session 3, GI infrastructure and use of spatial database Statistics Finland, Population
enzymatic families regulated in response to a short-term cytokinin treatment in root apices
 A B C D Supplemental tlfigure 1. 1Mtbli Metabolic processes, hormonal pathways and dlarge enzymatic families regulated in response to a short-term cytokinin treatment in root apices The 297 transcripts
A B C D Supplemental tlfigure 1. 1Mtbli Metabolic processes, hormonal pathways and dlarge enzymatic families regulated in response to a short-term cytokinin treatment in root apices The 297 transcripts
Tree map system in harvester
 Tree map system in harvester Fibic seminar 12.6.2013 Lahti Timo Melkas, Metsäteho Oy Mikko Miettinen, Argone Oy Kalle Einola, Ponsse Oyj Project goals EffFibre project 2011-2013 (WP3) To evaluate the accuracy
Tree map system in harvester Fibic seminar 12.6.2013 Lahti Timo Melkas, Metsäteho Oy Mikko Miettinen, Argone Oy Kalle Einola, Ponsse Oyj Project goals EffFibre project 2011-2013 (WP3) To evaluate the accuracy
The CCR Model and Production Correspondence
 The CCR Model and Production Correspondence Tim Schöneberg The 19th of September Agenda Introduction Definitions Production Possiblity Set CCR Model and the Dual Problem Input excesses and output shortfalls
The CCR Model and Production Correspondence Tim Schöneberg The 19th of September Agenda Introduction Definitions Production Possiblity Set CCR Model and the Dual Problem Input excesses and output shortfalls
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table 5.11.2013 16:44 / 1 Minimum
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table 5.11.2013 16:44 / 1 Minimum
TEST REPORT Nro VTT-S Air tightness and strength tests for Furanflex exhaust air ducts
 TEST REPORT Nro VTT-S-04515-08 19.5.008 Air tightness and strength tests for Furanflex exhaust air ducts Requested by: Hormex Oy TEST REPORT NRO VTT-S-04515-08 1 () Requested by Order Hormex Oy Linnanherrankuja
TEST REPORT Nro VTT-S-04515-08 19.5.008 Air tightness and strength tests for Furanflex exhaust air ducts Requested by: Hormex Oy TEST REPORT NRO VTT-S-04515-08 1 () Requested by Order Hormex Oy Linnanherrankuja
HARJOITUS- PAKETTI A
 Logistiikka A35A00310 Tuotantotalouden perusteet HARJOITUS- PAKETTI A (6 pistettä) TUTA 19 Luento 3.Ennustaminen County General 1 piste The number of heart surgeries performed at County General Hospital
Logistiikka A35A00310 Tuotantotalouden perusteet HARJOITUS- PAKETTI A (6 pistettä) TUTA 19 Luento 3.Ennustaminen County General 1 piste The number of heart surgeries performed at County General Hospital
mtorc1 and CK2 coordinate ternary and eif4f complex assembly
 mtorc1 and CK2 coordinate ternary and eif4f complex assembly Valentina Gandin 1,2,3,4 *, Laia Masvidal 5 *, Marie Cargnello 1,2,3,4 *, Laszlo Gyenis 6, Shannon McLaughlan 1,2,3,4, Yutian Cai 1,2,3,4, Clara
mtorc1 and CK2 coordinate ternary and eif4f complex assembly Valentina Gandin 1,2,3,4 *, Laia Masvidal 5 *, Marie Cargnello 1,2,3,4 *, Laszlo Gyenis 6, Shannon McLaughlan 1,2,3,4, Yutian Cai 1,2,3,4, Clara
Rakennukset Varjostus "real case" h/a 0,5 1,5
 Tuulivoimala Rakennukset Asuinrakennus Liikerak. tai Julkinen rak. Lomarakennus Teollinen rakennus Kirkollinen rakennus Varjostus "real case" h/a 1 h/a 8 h/a 20 h/a 1 2 3 5 8 4 6 7 9 10 0 0,5 1 1,5 2 km
Tuulivoimala Rakennukset Asuinrakennus Liikerak. tai Julkinen rak. Lomarakennus Teollinen rakennus Kirkollinen rakennus Varjostus "real case" h/a 1 h/a 8 h/a 20 h/a 1 2 3 5 8 4 6 7 9 10 0 0,5 1 1,5 2 km
Supplementary Information
 Supplementary Information Supplementary Table 1. Strain table Strain name Genotype Reference CEN.PK111-27B MATa leu2 trp1 Euroscarf TC-49 TC-50 pdc5::aro4* aro10::aro7* This study TC-50 MATa leu2 trp1
Supplementary Information Supplementary Table 1. Strain table Strain name Genotype Reference CEN.PK111-27B MATa leu2 trp1 Euroscarf TC-49 TC-50 pdc5::aro4* aro10::aro7* This study TC-50 MATa leu2 trp1
I. Principles of Pointer Year Analysis
 I. Principles of Pointer Year Analysis Fig 1. Maximum (red) and minimum (blue) pointer years. 1 Fig 2. Principle of pointer year calculation. Fig 3. Skeleton plot graph created by Kinsys/Kigraph programme.
I. Principles of Pointer Year Analysis Fig 1. Maximum (red) and minimum (blue) pointer years. 1 Fig 2. Principle of pointer year calculation. Fig 3. Skeleton plot graph created by Kinsys/Kigraph programme.
LYTH-CONS CONSISTENCY TRANSMITTER
 LYTH-CONS CONSISTENCY TRANSMITTER LYTH-INSTRUMENT OY has generate new consistency transmitter with blade-system to meet high technical requirements in Pulp&Paper industries. Insurmountable advantages are
LYTH-CONS CONSISTENCY TRANSMITTER LYTH-INSTRUMENT OY has generate new consistency transmitter with blade-system to meet high technical requirements in Pulp&Paper industries. Insurmountable advantages are
13.11. Tulosten arviointi. tulosten arviointi. voimmeko luottaa saamiimme tuloksiin?
 13.11. tulosten arviointi Tulosten arviointi voimmeko luottaa saamiimme tuloksiin? onko osa saaduista tuloksista sattumanvaraisia? mitkä OSAT puusta ovat luotettavimpia? 1 KONSENSUSDIAGRAMMI Useita yhtä
13.11. tulosten arviointi Tulosten arviointi voimmeko luottaa saamiimme tuloksiin? onko osa saaduista tuloksista sattumanvaraisia? mitkä OSAT puusta ovat luotettavimpia? 1 KONSENSUSDIAGRAMMI Useita yhtä
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
TM ETRS-TM35FIN-ETRS89 WTG
 SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
SHADOW - Main Result Assumptions for shadow calculations Maximum distance for influence Calculate only when more than 20 % of sun is covered by the blade Please look in WTG table WindPRO version 2.8.579
Bioinformatics. Sequence Analysis: Part III. Pattern Searching and Gene Finding. Fran Lewitter, Ph.D. Head, Biocomputing Whitehead Institute
 Bioinformatics Sequence Analysis: Part III. Pattern Searching and Gene Finding Fran Lewitter, Ph.D. Head, Biocomputing Whitehead Institute Course Syllabus Jan 7 Jan 14 Jan 21 Jan 28 Feb 4 Feb 11 Feb 18
Bioinformatics Sequence Analysis: Part III. Pattern Searching and Gene Finding Fran Lewitter, Ph.D. Head, Biocomputing Whitehead Institute Course Syllabus Jan 7 Jan 14 Jan 21 Jan 28 Feb 4 Feb 11 Feb 18
Genome 373: Genomic Informatics. Professors Elhanan Borenstein and Jay Shendure
 Genome 373: Genomic Informatics Professors Elhanan Borenstein and Jay Shendure Genome 373 This course is intended to introduce students to the breadth of problems and methods in computational analysis
Genome 373: Genomic Informatics Professors Elhanan Borenstein and Jay Shendure Genome 373 This course is intended to introduce students to the breadth of problems and methods in computational analysis
Predicting evolutionarily conserved regions (ECRs) in the Xenopus tropicalis genome using a MultiPipMaker-based bioinformatic strategy
 Predicting evolutionarily conserved regions (ECRs) in the Xenopus tropicalis genome using a MultiPipMaker-based bioinformatic strategy OVERVIEW- Method updated and written by Sarah Louie (6/19/08) Initially
Predicting evolutionarily conserved regions (ECRs) in the Xenopus tropicalis genome using a MultiPipMaker-based bioinformatic strategy OVERVIEW- Method updated and written by Sarah Louie (6/19/08) Initially
Plant protection of cereals current situation
 Plant protection of cereals current situation PesticideLife Opening seminar 19.2.2010, Jokioinen Pertti Rajala Cereals, total 1 203,1 Winter wheat 16,4 Spring wheat 201,9 Winter rye 11,3 Sring rye 5,1
Plant protection of cereals current situation PesticideLife Opening seminar 19.2.2010, Jokioinen Pertti Rajala Cereals, total 1 203,1 Winter wheat 16,4 Spring wheat 201,9 Winter rye 11,3 Sring rye 5,1
Constructive Alignment in Specialisation Studies in Industrial Pharmacy in Finland
 Constructive Alignment in Specialisation Studies in Industrial Pharmacy in Finland Anne Mari Juppo, Nina Katajavuori University of Helsinki Faculty of Pharmacy 23.7.2012 1 Background Pedagogic research
Constructive Alignment in Specialisation Studies in Industrial Pharmacy in Finland Anne Mari Juppo, Nina Katajavuori University of Helsinki Faculty of Pharmacy 23.7.2012 1 Background Pedagogic research
VIIKKI BIOCENTER University of Helsinki
 VIIKKI BIOCENTER University of Helsinki Mitä uudet DNA sekvensointimenetelmät voivat paljastaa luonnonjärjestelmästä? Petri Auvinen DNA Sequencing and Genomics Laboratory Institute of Biotechnology Petri
VIIKKI BIOCENTER University of Helsinki Mitä uudet DNA sekvensointimenetelmät voivat paljastaa luonnonjärjestelmästä? Petri Auvinen DNA Sequencing and Genomics Laboratory Institute of Biotechnology Petri
WAMS 2010,Ylivieska Monitoring service of energy efficiency in housing. 13.10.2010 Jan Nyman, jan.nyman@posintra.fi
 WAMS 2010,Ylivieska Monitoring service of energy efficiency in housing 13.10.2010 Jan Nyman, jan.nyman@posintra.fi Background info STOK: development center for technology related to building automation
WAMS 2010,Ylivieska Monitoring service of energy efficiency in housing 13.10.2010 Jan Nyman, jan.nyman@posintra.fi Background info STOK: development center for technology related to building automation
 Digitally signed by Hans Vadbäck DN: cn=hans Vadbäck, o, ou=fcg Suunnittelu ja Tekniikka Oy, email=hans.vadback@fcg.fi, c=fi Date: 2016.12.20 15:45:35 +02'00' Jakob Kjellman Digitally signed by Jakob Kjellman
Digitally signed by Hans Vadbäck DN: cn=hans Vadbäck, o, ou=fcg Suunnittelu ja Tekniikka Oy, email=hans.vadback@fcg.fi, c=fi Date: 2016.12.20 15:45:35 +02'00' Jakob Kjellman Digitally signed by Jakob Kjellman
3 9-VUOTIAIDEN LASTEN SUORIUTUMINEN BOSTONIN NIMENTÄTESTISTÄ
 Puhe ja kieli, 27:4, 141 147 (2007) 3 9-VUOTIAIDEN LASTEN SUORIUTUMINEN BOSTONIN NIMENTÄTESTISTÄ Soile Loukusa, Oulun yliopisto, suomen kielen, informaatiotutkimuksen ja logopedian laitos & University
Puhe ja kieli, 27:4, 141 147 (2007) 3 9-VUOTIAIDEN LASTEN SUORIUTUMINEN BOSTONIN NIMENTÄTESTISTÄ Soile Loukusa, Oulun yliopisto, suomen kielen, informaatiotutkimuksen ja logopedian laitos & University
Viral DNA as a model for coil to globule transition
 Viral DNA as a model for coil to globule transition Marina Rossi Lab. of complex fluids and molecular biophysics LITA (Segrate) UNIVERSITA DEGLI STUDI DI MILANO - PhD Workshop October 14 th, 2013 Temperature
Viral DNA as a model for coil to globule transition Marina Rossi Lab. of complex fluids and molecular biophysics LITA (Segrate) UNIVERSITA DEGLI STUDI DI MILANO - PhD Workshop October 14 th, 2013 Temperature
Nordea Bank Abp. Nordea Bank Abp
 Notice Nordea Bank Abp ISSUANCE by Nordea Bank Abp of instruments listed in Annex 1 Issued pursuant to the Structured Note Programme (the Notes ) Credit Event Notice On 17/12/2018 ISDA s (International
Notice Nordea Bank Abp ISSUANCE by Nordea Bank Abp of instruments listed in Annex 1 Issued pursuant to the Structured Note Programme (the Notes ) Credit Event Notice On 17/12/2018 ISDA s (International
ReFuel 70 % Emission Reduction Using Renewable High Cetane Number Paraffinic Diesel Fuel. Kalle Lehto, Aalto-yliopisto 5.5.
 ReFuel 70 % Emission Reduction Using Renewable High Cetane Number Paraffinic Diesel Fuel Kalle Lehto, Aalto-yliopisto 5.5.2011 Otaniemi ReFuel a three year research project (2009-2011) goal utilize the
ReFuel 70 % Emission Reduction Using Renewable High Cetane Number Paraffinic Diesel Fuel Kalle Lehto, Aalto-yliopisto 5.5.2011 Otaniemi ReFuel a three year research project (2009-2011) goal utilize the
OMINAISUUDET SOVELLUS. Technical data sheet C2 - C4 - BULLDOG. Bulldogpuunsitojalevyt
 Bulldog-puunsitojalevyjen avulla vahvistetaan puurakenteiden pulttiliitoksia. Kaksipuolisia puunsitojalevyjä käytetään vain puu-puuliitoksissa, mutta yksipuolisia puunsitojalevyjä voidaan käyttää myös
Bulldog-puunsitojalevyjen avulla vahvistetaan puurakenteiden pulttiliitoksia. Kaksipuolisia puunsitojalevyjä käytetään vain puu-puuliitoksissa, mutta yksipuolisia puunsitojalevyjä voidaan käyttää myös
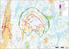
( N117 x HH141 ( Honkajoki N117 x 9 x HH120 tv-alueet ( ( ( ( ( ( ( ( ( ( m. Honkajoki & Kankaanpää tuulivoimahankkeet
 Honkajoki & Kankaanpää tuulivoimahankkeet N117 x HH141 Honkajoki N117 x 9 x HH120 tv-alueet Alahonkajoki_kaava_alueen_raja_polyline Asuinrakennus Julkinen tai liiker rak. Lomarakennus Teollinen rak. Allas
Honkajoki & Kankaanpää tuulivoimahankkeet N117 x HH141 Honkajoki N117 x 9 x HH120 tv-alueet Alahonkajoki_kaava_alueen_raja_polyline Asuinrakennus Julkinen tai liiker rak. Lomarakennus Teollinen rak. Allas
Supplemental Data. Manavski et al. (2015). Plant Cell /tpc
 1 A Promoter Transcription start Physcomitrella patens TGTGTAATAATAAGTTTTAATACCTGCCCCAGAG-AACTTCTCTATCATAGTCGTTCCGGTTCTAAAC---TGATCGAAA------------------------AGGAGT-- Marchantia polymorpha TGTGTCATAATAAATTTGAAGACCTGCCT-AGGT-AATTTCTGTATAATAGTTGTTCCAGTTTTAAAC---TGA--GAAA------------------------GAGATCTA
1 A Promoter Transcription start Physcomitrella patens TGTGTAATAATAAGTTTTAATACCTGCCCCAGAG-AACTTCTCTATCATAGTCGTTCCGGTTCTAAAC---TGATCGAAA------------------------AGGAGT-- Marchantia polymorpha TGTGTCATAATAAATTTGAAGACCTGCCT-AGGT-AATTTCTGTATAATAGTTGTTCCAGTTTTAAAC---TGA--GAAA------------------------GAGATCTA
Supporting Information for Electrophoresis DOI /elps Sunil Archak, Vemireddy Lakshminarayanareddy and Javaregowda Nagaraju
 ELECTROPHORESIS Supporting Information for Electrophoresis DOI 10.1002/elps.200600646 Sunil Archak, Vemireddy Lakshminarayanareddy and Javaregowda Nagaraju High-throughput multiplex microsatellite marker
ELECTROPHORESIS Supporting Information for Electrophoresis DOI 10.1002/elps.200600646 Sunil Archak, Vemireddy Lakshminarayanareddy and Javaregowda Nagaraju High-throughput multiplex microsatellite marker
Title: Enhancement of protein production via the strong DIT1 terminator and two RNA-binding proteins in Saccharomyces cerevisiae
 Title: Enhancement of protein production via the strong DIT1 terminator and two RNA-binding proteins in Saccharomyces cerevisiae Authors: Yoichiro Ito, Takao Kitagawa, Mamoru Yamanishi, Satoshi Katahira,
Title: Enhancement of protein production via the strong DIT1 terminator and two RNA-binding proteins in Saccharomyces cerevisiae Authors: Yoichiro Ito, Takao Kitagawa, Mamoru Yamanishi, Satoshi Katahira,
Research plan for masters thesis in forest sciences. The PELLETime 2009 Symposium Mervi Juntunen
 Modelling tree and stand characteristics and estimating biomass removals and harvesting costs of lodgepole pine (Pinus contorta) plantations in Iceland Research plan for masters thesis in forest sciences
Modelling tree and stand characteristics and estimating biomass removals and harvesting costs of lodgepole pine (Pinus contorta) plantations in Iceland Research plan for masters thesis in forest sciences
Chapter 7. Motif finding (week 11) Chapter 8. Sequence binning (week 11)
 Course organization Introduction ( Week 1) Part I: Algorithms for Sequence Analysis (Week 1-11) Chapter 1-3, Models and theories» Probability theory and Statistics (Week 2)» Algorithm complexity analysis
Course organization Introduction ( Week 1) Part I: Algorithms for Sequence Analysis (Week 1-11) Chapter 1-3, Models and theories» Probability theory and Statistics (Week 2)» Algorithm complexity analysis
Python Libraries 1 / 14
 Python Libraries 1 / 14 Gzip, CSV https://docs.python.org/3/library/gzip.html https://docs.python.org/3/library/csv.html https://daler.github.io/pybedtools/ - PyBed tools http://biopython.org/wiki/gff_parsing
Python Libraries 1 / 14 Gzip, CSV https://docs.python.org/3/library/gzip.html https://docs.python.org/3/library/csv.html https://daler.github.io/pybedtools/ - PyBed tools http://biopython.org/wiki/gff_parsing
Curriculum. Gym card
 A new school year Curriculum Fast Track Final Grading Gym card TET A new school year Work Ethic Detention Own work Organisation and independence Wilma TMU Support Services Well-Being CURRICULUM FAST TRACK
A new school year Curriculum Fast Track Final Grading Gym card TET A new school year Work Ethic Detention Own work Organisation and independence Wilma TMU Support Services Well-Being CURRICULUM FAST TRACK
Supporting Information Table S1 Core regions of 18 identified promoters in C. acetobutylicum
 Supporting Information Table S1 Core regions of 18 identified promoters in C. acetobutylicum Gene Core regions of the promoters Reference 35 region 10 region thl TATA TTGATA AAAATAATAATAGTGGG TATAAT TAA
Supporting Information Table S1 Core regions of 18 identified promoters in C. acetobutylicum Gene Core regions of the promoters Reference 35 region 10 region thl TATA TTGATA AAAATAATAATAGTGGG TATAAT TAA
Nordea Bank Abp. Nordea Bank Abp
 Notice Nordea Bank Abp ISSUANCE by Nordea Bank Abp of instruments listed in Annex 1 Issued pursuant to the Structured Note Programme (the Notes ) Credit Event Notice On 15/10/2018 ISDA s (International
Notice Nordea Bank Abp ISSUANCE by Nordea Bank Abp of instruments listed in Annex 1 Issued pursuant to the Structured Note Programme (the Notes ) Credit Event Notice On 15/10/2018 ISDA s (International
Social and Regional Economic Impacts of Use of Bioenergy and Energy Wood Harvesting in Suomussalmi
 Social and Regional Economic Impacts of Use of Bioenergy and Energy Wood Harvesting in Suomussalmi Green Cities and Settlements 18.2.2014 Ville Manninen Writers Project group Sirpa Korhonen, Anna Mari
Social and Regional Economic Impacts of Use of Bioenergy and Energy Wood Harvesting in Suomussalmi Green Cities and Settlements 18.2.2014 Ville Manninen Writers Project group Sirpa Korhonen, Anna Mari
Results on the new polydrug use questions in the Finnish TDI data
 Results on the new polydrug use questions in the Finnish TDI data Multi-drug use, polydrug use and problematic polydrug use Martta Forsell, Finnish Focal Point 28/09/2015 Martta Forsell 1 28/09/2015 Esityksen
Results on the new polydrug use questions in the Finnish TDI data Multi-drug use, polydrug use and problematic polydrug use Martta Forsell, Finnish Focal Point 28/09/2015 Martta Forsell 1 28/09/2015 Esityksen
Information on preparing Presentation
 Information on preparing Presentation Seminar on big data management Lecturer: Spring 2017 20.1.2017 1 Agenda Hints and tips on giving a good presentation Watch two videos and discussion 22.1.2017 2 Goals
Information on preparing Presentation Seminar on big data management Lecturer: Spring 2017 20.1.2017 1 Agenda Hints and tips on giving a good presentation Watch two videos and discussion 22.1.2017 2 Goals
EU GMP Guide Part IV: Guideline on GMP spesific to ATMP. Pirjo Hänninen
 EU GMP Guide Part IV: Guideline on GMP spesific to ATMP Pirjo Hänninen 3.4.2019 ATMP ATMPs: Gene therapy medicinal products Somatic cell therapy medicinal products Tissue engineered products Vuosi-kk-pv
EU GMP Guide Part IV: Guideline on GMP spesific to ATMP Pirjo Hänninen 3.4.2019 ATMP ATMPs: Gene therapy medicinal products Somatic cell therapy medicinal products Tissue engineered products Vuosi-kk-pv
Lausuntopyyntöluettelo HUOM. Komiteoiden ja seurantaryhmien kokoonpanot on esitetty SESKOn komitealuettelossa
 1(11) pren ISO 15223-1 rev Medical devices - Symbols to be used with medical device labels, labelling and information to be supplied - Part 1: General requirements Kansainvälinen valmisteluvaihe: 90/385/EEC,
1(11) pren ISO 15223-1 rev Medical devices - Symbols to be used with medical device labels, labelling and information to be supplied - Part 1: General requirements Kansainvälinen valmisteluvaihe: 90/385/EEC,
CONVERSION KITS FOR FRAMES MR8 AND MR9 INSTALLATION INSTRUCTIONS
 CONVERSION KITS FOR FRAMES MR8 AND MR9 INSTALLATION INSTRUCTIONS MR8 AND MR9 CONVERSION KIT - INSTALLATION INSTRUCTION Document ID: DPD01787, Revision: A, Release date: 17.11.2015 1.1 MR8 kit contents
CONVERSION KITS FOR FRAMES MR8 AND MR9 INSTALLATION INSTRUCTIONS MR8 AND MR9 CONVERSION KIT - INSTALLATION INSTRUCTION Document ID: DPD01787, Revision: A, Release date: 17.11.2015 1.1 MR8 kit contents
LUONNOS RT 80260 EN AGREEMENT ON BUILDING WORKS 1 THE PARTIES. May 1998 1 (10)
 RT 80260 EN May 1998 1 (10) AGREEMENT ON BUILDING WORKS This agreement template is based on the General Terms and Conditions of Building Contracts YSE 1998 RT 16-10660, LVI 03-10277, Ratu 417-7, KH X4-00241.
RT 80260 EN May 1998 1 (10) AGREEMENT ON BUILDING WORKS This agreement template is based on the General Terms and Conditions of Building Contracts YSE 1998 RT 16-10660, LVI 03-10277, Ratu 417-7, KH X4-00241.
Telecommunication Software
 Telecommunication Software Final exam 21.11.2006 COMPUTER ENGINEERING LABORATORY 521265A Vastaukset englanniksi tai suomeksi. / Answers in English or in Finnish. 1. (a) Määrittele sovellusviesti, PersonnelRecord,
Telecommunication Software Final exam 21.11.2006 COMPUTER ENGINEERING LABORATORY 521265A Vastaukset englanniksi tai suomeksi. / Answers in English or in Finnish. 1. (a) Määrittele sovellusviesti, PersonnelRecord,
Särmäystyökalut kuvasto Press brake tools catalogue
 Finnish sheet metal machinery know-how since 1978 Särmäystyökalut kuvasto Press brake tools catalogue www.aliko.fi ALIKO bending chart Required capacity in kn (T) in relation to V-opening. V R A S = plates
Finnish sheet metal machinery know-how since 1978 Särmäystyökalut kuvasto Press brake tools catalogue www.aliko.fi ALIKO bending chart Required capacity in kn (T) in relation to V-opening. V R A S = plates
Characterization of clay using x-ray and neutron scattering at the University of Helsinki and ILL
 Characterization of clay using x-ray and neutron scattering at the University of Helsinki and ILL Ville Liljeström, Micha Matusewicz, Kari Pirkkalainen, Jussi-Petteri Suuronen and Ritva Serimaa 13.3.2012
Characterization of clay using x-ray and neutron scattering at the University of Helsinki and ILL Ville Liljeström, Micha Matusewicz, Kari Pirkkalainen, Jussi-Petteri Suuronen and Ritva Serimaa 13.3.2012
SEMINAARI SFS:SSÄ UUDET YHTEISET STANDARDIT YMPÄRISTÖANALYTIIKKAAN? 13.5.2014 PENTTI MANNINEN
 SEMINAARI SFS:SSÄ UUDET YHTEISET STANDARDIT YMPÄRISTÖANALYTIIKKAAN? 13.5.2014 PENTTI MANNINEN PROJECT HORIZONTAL (1) Euroopan komission rahoittama hanke, jonka puitteissa oli tarkoitus luoda yhtenäisiä
SEMINAARI SFS:SSÄ UUDET YHTEISET STANDARDIT YMPÄRISTÖANALYTIIKKAAN? 13.5.2014 PENTTI MANNINEN PROJECT HORIZONTAL (1) Euroopan komission rahoittama hanke, jonka puitteissa oli tarkoitus luoda yhtenäisiä
AKKREDITOITU TESTAUSLABORATORIO ACCREDITED TESTING LABORATORY VERKOTAN OY VERKOTAN LTD.
 T287/M03/2017 Liite 1 / Appendix 1 Sivu / Page 1(5) AKKREDITOITU TESTAUSLABORATORIO ACCREDITED TESTING LABORATORY VERKOTAN OY VERKOTAN LTD. Tunnus Code Laboratorio Laboratory Osoite Address www www T287
T287/M03/2017 Liite 1 / Appendix 1 Sivu / Page 1(5) AKKREDITOITU TESTAUSLABORATORIO ACCREDITED TESTING LABORATORY VERKOTAN OY VERKOTAN LTD. Tunnus Code Laboratorio Laboratory Osoite Address www www T287
Paikkatiedon semanttinen mallinnus, integrointi ja julkaiseminen Case Suomalainen ajallinen paikkaontologia SAPO
 Paikkatiedon semanttinen mallinnus, integrointi ja julkaiseminen Case Suomalainen ajallinen paikkaontologia SAPO Tomi Kauppinen, Eero Hyvönen, Jari Väätäinen Semantic Computing Research Group (SeCo) http://www.seco.tkk.fi/
Paikkatiedon semanttinen mallinnus, integrointi ja julkaiseminen Case Suomalainen ajallinen paikkaontologia SAPO Tomi Kauppinen, Eero Hyvönen, Jari Väätäinen Semantic Computing Research Group (SeCo) http://www.seco.tkk.fi/
Choose Finland-Helsinki Valitse Finland-Helsinki
 Write down the Temporary Application ID. If you do not manage to complete the form you can continue where you stopped with this ID no. Muista Temporary Application ID. Jos et onnistu täyttää lomake loppuun
Write down the Temporary Application ID. If you do not manage to complete the form you can continue where you stopped with this ID no. Muista Temporary Application ID. Jos et onnistu täyttää lomake loppuun
Exercise 1. (session: )
 EEN-E3001, FUNDAMENTALS IN INDUSTRIAL ENERGY ENGINEERING Exercise 1 (session: 24.1.2017) Problem 3 will be graded. The deadline for the return is on 31.1. at 12:00 am (before the exercise session). You
EEN-E3001, FUNDAMENTALS IN INDUSTRIAL ENERGY ENGINEERING Exercise 1 (session: 24.1.2017) Problem 3 will be graded. The deadline for the return is on 31.1. at 12:00 am (before the exercise session). You
